Please be patient as the page loads
|
ST5_HUMAN
|
||||||
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DEN2A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.963604 (rank : 6) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
DEN2A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.967392 (rank : 3) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
DEN2C_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.964497 (rank : 5) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q68D51, Q5TCX6, Q6P3R3 | Gene names | DENND2C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
DEN2C_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.965384 (rank : 4) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P9P8, Q3T9X2, Q3UV30, Q80V46 | Gene names | Dennd2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
ST5_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 151 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
ST5_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.985741 (rank : 2) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
DEN2D_HUMAN
|
||||||
| θ value | 8.99321e-117 (rank : 7) | NC score | 0.956434 (rank : 8) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H6A0, Q5T5V6, Q9BSU0 | Gene names | DENND2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
DEN2D_MOUSE
|
||||||
| θ value | 1.29861e-115 (rank : 8) | NC score | 0.956818 (rank : 7) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91VV4, Q3U028, Q9D831 | Gene names | Dennd2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
DEN1A_MOUSE
|
||||||
| θ value | 2.42216e-37 (rank : 9) | NC score | 0.788595 (rank : 9) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DEN1A_HUMAN
|
||||||
| θ value | 1.20211e-36 (rank : 10) | NC score | 0.782756 (rank : 10) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
RA6I1_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 11) | NC score | 0.559481 (rank : 12) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 12) | NC score | 0.560149 (rank : 11) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
MYCPP_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 13) | NC score | 0.556571 (rank : 13) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z401, Q14655, Q86T77, Q8IVX2, Q8NB93 | Gene names | MYCPBP, IRLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-myc promoter-binding protein. | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 14) | NC score | 0.412116 (rank : 15) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
MTMRD_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 15) | NC score | 0.411494 (rank : 16) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
DEN4C_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 16) | NC score | 0.533849 (rank : 14) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5VZ89, Q6AI48, Q6ZUB3, Q8NCY7, Q9H6N4, Q9NUT1, Q9NWA5 | Gene names | DENND4C, C9orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 4C. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 17) | NC score | 0.057894 (rank : 22) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 18) | NC score | 0.068089 (rank : 21) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 19) | NC score | 0.073548 (rank : 20) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
DACT1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.083030 (rank : 19) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 21) | NC score | 0.047840 (rank : 28) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 22) | NC score | 0.085227 (rank : 17) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.084930 (rank : 18) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.030491 (rank : 56) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.051313 (rank : 27) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.028075 (rank : 66) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.053382 (rank : 25) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.017031 (rank : 94) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.036118 (rank : 41) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 0.125558 (rank : 30) | NC score | 0.045170 (rank : 30) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 31) | NC score | 0.024726 (rank : 79) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.013749 (rank : 102) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.035719 (rank : 43) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 34) | NC score | 0.046398 (rank : 29) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RBMX2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.032228 (rank : 53) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |
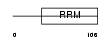
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 36) | NC score | 0.008009 (rank : 125) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
B4GN3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.030353 (rank : 57) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.044644 (rank : 32) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.052732 (rank : 26) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
BOC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.006985 (rank : 128) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6AZB0, Q6KAM5, Q6P5H3, Q7TMJ3, Q8CE73, Q8CE91, Q8R377, Q923W7 | Gene names | Boc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
DC1L2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.033414 (rank : 46) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.040397 (rank : 36) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.031796 (rank : 54) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.025897 (rank : 75) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | -0.000088 (rank : 149) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.029693 (rank : 59) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.002320 (rank : 144) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
DUT_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.041358 (rank : 35) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33316, O14785, Q16708, Q16860, Q96Q81 | Gene names | DUT | |||
|
Domain Architecture |

|
|||||
| Description | Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial precursor (EC 3.6.1.23) (dUTPase) (dUTP pyrophosphatase). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.043531 (rank : 33) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
NFIC_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.019070 (rank : 92) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70255, O09072, P70256, Q3U2I9, Q99MA3, Q99MA4, Q99MA5, Q99MA6, Q99MA7, Q99MA8, Q9R1G3 | Gene names | Nfic | |||
|
Domain Architecture |
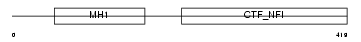
|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.026414 (rank : 72) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
TIP60_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.025785 (rank : 76) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92993, O95624, Q13430, Q6GSE8, Q9BWK7 | Gene names | HTATIP, TIP60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat interactive protein) (cPLA(2)-interacting protein). | |||||
|
ASXL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.032674 (rank : 52) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
GP152_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.020140 (rank : 89) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.028725 (rank : 62) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
RASF7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.024531 (rank : 80) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DD19 | Gene names | Rassf7 | |||
|
Domain Architecture |
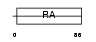
|
|||||
| Description | Ras association domain-containing protein 7. | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.033023 (rank : 48) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.032732 (rank : 51) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PCIF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.029780 (rank : 58) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.033236 (rank : 47) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RBNS5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.028357 (rank : 64) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.014568 (rank : 99) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.056337 (rank : 23) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TIP60_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.024753 (rank : 78) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.008015 (rank : 124) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.013018 (rank : 104) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
GTSE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.041700 (rank : 34) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
KITH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.024771 (rank : 77) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04183, Q969V0, Q9UMG9 | Gene names | TK1 | |||
|
Domain Architecture |
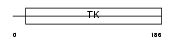
|
|||||
| Description | Thymidine kinase, cytosolic (EC 2.7.1.21). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.035426 (rank : 44) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.033011 (rank : 49) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.039470 (rank : 37) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.036240 (rank : 39) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.029026 (rank : 60) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.034186 (rank : 45) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.045092 (rank : 31) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MICA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.008166 (rank : 121) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | -0.000740 (rank : 152) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SPF45_MOUSE
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.021942 (rank : 83) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.036082 (rank : 42) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.026104 (rank : 74) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.014410 (rank : 100) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.021744 (rank : 84) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.028034 (rank : 68) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
NOP56_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.017891 (rank : 93) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
SEM6A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.004815 (rank : 138) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
TNR4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.013014 (rank : 105) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |
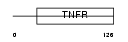
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
CENPC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.026607 (rank : 71) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
EVI2B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.020776 (rank : 87) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P34910 | Gene names | EVI2B, EVDB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.016570 (rank : 95) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PIGQ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.015815 (rank : 96) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.031729 (rank : 55) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.013370 (rank : 103) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
STOX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.032789 (rank : 50) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.028186 (rank : 65) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CT160_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.013875 (rank : 101) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUG4, Q5JYR9, Q8N5F1, Q8N6G8, Q96MD5 | Gene names | C20orf160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf160. | |||||
|
INVS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.005045 (rank : 137) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
RBBP8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.012294 (rank : 109) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
RIMB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.009341 (rank : 116) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
SI1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.007305 (rank : 126) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SLIK3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.002899 (rank : 142) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94933 | Gene names | SLITRK3, KIAA0848 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLIT and NTRK-like protein 3 precursor. | |||||
|
SON_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.036901 (rank : 38) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.028070 (rank : 67) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
WDHD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.006616 (rank : 132) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.009919 (rank : 115) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
YTHD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.015573 (rank : 97) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y5A9, Q5VSZ9, Q8TDH0, Q9BUJ5 | Gene names | YTHDF2, HGRG8 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 2 (High-glucose-regulated protein 8) (NY- REN-2 antigen) (CLL-associated antigen KW-14). | |||||
|
ATRIP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.010599 (rank : 113) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BMG1 | Gene names | Atrip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
CP2J5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.001503 (rank : 146) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54749 | Gene names | Cyp2j5 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2J5 (EC 1.14.14.1) (CYPIIJ5) (Arachidonic acid epoxygenase). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.008033 (rank : 123) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
G3BP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.012114 (rank : 110) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97855 | Gene names | G3bp, G3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.020083 (rank : 90) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.021145 (rank : 85) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.004785 (rank : 139) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.022819 (rank : 82) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.000092 (rank : 148) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
ZO2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.006817 (rank : 130) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
AT1A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.005311 (rank : 135) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.005305 (rank : 136) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
CF152_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.020155 (rank : 88) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.005623 (rank : 134) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
GAB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.015509 (rank : 98) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.007275 (rank : 127) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.006689 (rank : 131) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
LRBA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.006850 (rank : 129) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.008747 (rank : 120) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.023077 (rank : 81) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.004471 (rank : 140) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PHF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.008949 (rank : 117) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
RBFAL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.011992 (rank : 111) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P3B9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ribosome-binding factor A, mitochondrial precursor. | |||||
|
SHOC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.001779 (rank : 145) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ13, O76063 | Gene names | SHOC2, KIAA0862 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
SIA7A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.012319 (rank : 107) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.021017 (rank : 86) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.010057 (rank : 114) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
CO4A4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.008057 (rank : 122) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |
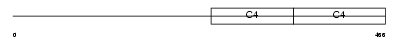
|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
DMRTD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.011826 (rank : 112) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.026391 (rank : 73) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.019555 (rank : 91) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
KI13A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.002651 (rank : 143) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.036206 (rank : 40) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | -0.000656 (rank : 151) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.028370 (rank : 63) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.027250 (rank : 69) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NPY4R_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | -0.000562 (rank : 150) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50391, Q13456 | Gene names | PPYR1, NPY4R | |||
|
Domain Architecture |
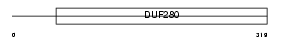
|
|||||
| Description | Neuropeptide Y receptor type 4 (NPY4-R) (Pancreatic polypeptide receptor 1) (PP1). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.003592 (rank : 141) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RT34_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.026665 (rank : 70) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P82930, Q9BVI7 | Gene names | MRPS34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 28S ribosomal protein S34 (S34mt) (MRP-S34). | |||||
|
SGIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.028752 (rank : 61) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SHOC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.001444 (rank : 147) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88520, Q3UJH6, Q8BVL0 | Gene names | Shoc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.008836 (rank : 118) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.006150 (rank : 133) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.012299 (rank : 108) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TRABD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.008822 (rank : 119) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4I3, Q96ED8, Q9H7N1, Q9UGX6, Q9UGX7 | Gene names | TRABD | |||
|
Domain Architecture |
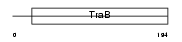
|
|||||
| Description | TraB domain-containing protein. | |||||
|
US6NL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.012968 (rank : 106) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.054228 (rank : 24) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ST5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 151 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
ST5_MOUSE
|
||||||
| NC score | 0.985741 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
DEN2A_MOUSE
|
||||||
| NC score | 0.967392 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
DEN2C_MOUSE
|
||||||
| NC score | 0.965384 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P9P8, Q3T9X2, Q3UV30, Q80V46 | Gene names | Dennd2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
DEN2C_HUMAN
|
||||||
| NC score | 0.964497 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q68D51, Q5TCX6, Q6P3R3 | Gene names | DENND2C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
DEN2A_HUMAN
|
||||||
| NC score | 0.963604 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
DEN2D_MOUSE
|
||||||
| NC score | 0.956818 (rank : 7) | θ value | 1.29861e-115 (rank : 8) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91VV4, Q3U028, Q9D831 | Gene names | Dennd2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
DEN2D_HUMAN
|
||||||
| NC score | 0.956434 (rank : 8) | θ value | 8.99321e-117 (rank : 7) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H6A0, Q5T5V6, Q9BSU0 | Gene names | DENND2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
DEN1A_MOUSE
|
||||||
| NC score | 0.788595 (rank : 9) | θ value | 2.42216e-37 (rank : 9) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DEN1A_HUMAN
|
||||||
| NC score | 0.782756 (rank : 10) | θ value | 1.20211e-36 (rank : 10) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.560149 (rank : 11) | θ value | 3.07829e-16 (rank : 12) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_HUMAN
|
||||||
| NC score | 0.559481 (rank : 12) | θ value | 3.07829e-16 (rank : 11) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
MYCPP_HUMAN
|
||||||
| NC score | 0.556571 (rank : 13) | θ value | 3.52202e-12 (rank : 13) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z401, Q14655, Q86T77, Q8IVX2, Q8NB93 | Gene names | MYCPBP, IRLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-myc promoter-binding protein. | |||||
|
DEN4C_HUMAN
|
||||||
| NC score | 0.533849 (rank : 14) | θ value | 1.25267e-09 (rank : 16) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5VZ89, Q6AI48, Q6ZUB3, Q8NCY7, Q9H6N4, Q9NUT1, Q9NWA5 | Gene names | DENND4C, C9orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 4C. | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.412116 (rank : 15) | θ value | 7.84624e-12 (rank : 14) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
MTMRD_HUMAN
|
||||||
| NC score | 0.411494 (rank : 16) | θ value | 2.98157e-11 (rank : 15) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.085227 (rank : 17) | θ value | 0.0252991 (rank : 22) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.084930 (rank : 18) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
DACT1_HUMAN
|
||||||
| NC score | 0.083030 (rank : 19) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.073548 (rank : 20) | θ value | 0.00228821 (rank : 19) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.068089 (rank : 21) | θ value | 0.00175202 (rank : 18) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.057894 (rank : 22) | θ value | 7.1131e-05 (rank : 17) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.056337 (rank : 23) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.054228 (rank : 24) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.053382 (rank : 25) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.052732 (rank : 26) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.051313 (rank : 27) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.047840 (rank : 28) | θ value | 0.0193708 (rank : 21) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.046398 (rank : 29) | θ value | 0.163984 (rank : 34) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.045170 (rank : 30) | θ value | 0.125558 (rank : 30) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.045092 (rank : 31) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.044644 (rank : 32) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.043531 (rank : 33) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
GTSE1_HUMAN
|
||||||
| NC score | 0.041700 (rank : 34) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
DUT_HUMAN
|
||||||
| NC score | 0.041358 (rank : 35) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33316, O14785, Q16708, Q16860, Q96Q81 | Gene names | DUT | |||
|
Domain Architecture |

|
|||||
| Description | Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial precursor (EC 3.6.1.23) (dUTPase) (dUTP pyrophosphatase). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.040397 (rank : 36) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.039470 (rank : 37) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.036901 (rank : 38) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.036240 (rank : 39) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.036206 (rank : 40) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.036118 (rank : 41) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.036082 (rank : 42) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.035719 (rank : 43) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.035426 (rank : 44) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.034186 (rank : 45) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
DC1L2_MOUSE
|
||||||
| NC score | 0.033414 (rank : 46) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.033236 (rank : 47) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.033023 (rank : 48) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.033011 (rank : 49) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
STOX1_HUMAN
|
||||||
| NC score | 0.032789 (rank : 50) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.032732 (rank : 51) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ASXL1_HUMAN
|
||||||
| NC score | 0.032674 (rank : 52) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
RBMX2_HUMAN
|
||||||
| NC score | 0.032228 (rank : 53) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.031796 (rank : 54) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.031729 (rank : 55) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.030491 (rank : 56) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
B4GN3_MOUSE
|
||||||
| NC score | 0.030353 (rank : 57) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
PCIF1_HUMAN
|
||||||
| NC score | 0.029780 (rank : 58) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.029693 (rank : 59) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.029026 (rank : 60) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SGIP1_HUMAN
|
||||||
| NC score | 0.028752 (rank : 61) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.028725 (rank : 62) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.028370 (rank : 63) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
RBNS5_HUMAN
|
||||||
| NC score | 0.028357 (rank : 64) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.028186 (rank : 65) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.028075 (rank : 66) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.028070 (rank : 67) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.028034 (rank : 68) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.027250 (rank : 69) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
RT34_HUMAN
|
||||||
| NC score | 0.026665 (rank : 70) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P82930, Q9BVI7 | Gene names | MRPS34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 28S ribosomal protein S34 (S34mt) (MRP-S34). | |||||
|
CENPC_HUMAN
|
||||||
| NC score | 0.026607 (rank : 71) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.026414 (rank : 72) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.026391 (rank : 73) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.026104 (rank : 74) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.025897 (rank : 75) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
TIP60_HUMAN
|
||||||
| NC score | 0.025785 (rank : 76) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92993, O95624, Q13430, Q6GSE8, Q9BWK7 | Gene names | HTATIP, TIP60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat interactive protein) (cPLA(2)-interacting protein). | |||||
|
KITH_HUMAN
|
||||||
| NC score | 0.024771 (rank : 77) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04183, Q969V0, Q9UMG9 | Gene names | TK1 | |||
|
Domain Architecture |

|
|||||
| Description | Thymidine kinase, cytosolic (EC 2.7.1.21). | |||||
|
TIP60_MOUSE
|
||||||
| NC score | 0.024753 (rank : 78) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.024726 (rank : 79) | θ value | 0.125558 (rank : 31) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
RASF7_MOUSE
|
||||||
| NC score | 0.024531 (rank : 80) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DD19 | Gene names | Rassf7 | |||
|
Domain Architecture |

|
|||||
| Description | Ras association domain-containing protein 7. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.023077 (rank : 81) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.022819 (rank : 82) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
SPF45_MOUSE
|
||||||
| NC score | 0.021942 (rank : 83) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.021744 (rank : 84) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.021145 (rank : 85) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.021017 (rank : 86) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
EVI2B_HUMAN
|
||||||
| NC score | 0.020776 (rank : 87) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P34910 | Gene names | EVI2B, EVDB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
CF152_MOUSE
|
||||||
| NC score | 0.020155 (rank : 88) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.020140 (rank : 89) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.020083 (rank : 90) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.019555 (rank : 91) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
NFIC_MOUSE
|
||||||
| NC score | 0.019070 (rank : 92) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70255, O09072, P70256, Q3U2I9, Q99MA3, Q99MA4, Q99MA5, Q99MA6, Q99MA7, Q99MA8, Q9R1G3 | Gene names | Nfic | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
NOP56_HUMAN
|
||||||
| NC score | 0.017891 (rank : 93) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.017031 (rank : 94) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.016570 (rank : 95) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PIGQ_HUMAN
|
||||||
| NC score | 0.015815 (rank : 96) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
YTHD2_HUMAN
|
||||||
| NC score | 0.015573 (rank : 97) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y5A9, Q5VSZ9, Q8TDH0, Q9BUJ5 | Gene names | YTHDF2, HGRG8 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 2 (High-glucose-regulated protein 8) (NY- REN-2 antigen) (CLL-associated antigen KW-14). | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.015509 (rank : 98) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.014568 (rank : 99) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.014410 (rank : 100) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
CT160_HUMAN
|
||||||
| NC score | 0.013875 (rank : 101) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUG4, Q5JYR9, Q8N5F1, Q8N6G8, Q96MD5 | Gene names | C20orf160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf160. | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.013749 (rank : 102) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.013370 (rank : 103) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.013018 (rank : 104) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
TNR4_HUMAN
|
||||||
| NC score | 0.013014 (rank : 105) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
US6NL_HUMAN
|
||||||
| NC score | 0.012968 (rank : 106) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
SIA7A_MOUSE
|
||||||
| NC score | 0.012319 (rank : 107) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.012299 (rank : 108) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RBBP8_HUMAN
|
||||||
| NC score | 0.012294 (rank : 109) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
G3BP_MOUSE
|
||||||
| NC score | 0.012114 (rank : 110) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97855 | Gene names | G3bp, G3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
RBFAL_MOUSE
|
||||||
| NC score | 0.011992 (rank : 111) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P3B9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ribosome-binding factor A, mitochondrial precursor. | |||||
|
DMRTD_HUMAN
|
||||||
| NC score | 0.011826 (rank : 112) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
ATRIP_MOUSE
|
||||||
| NC score | 0.010599 (rank : 113) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BMG1 | Gene names | Atrip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.010057 (rank : 114) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.009919 (rank : 115) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
RIMB2_HUMAN
|
||||||
| NC score | 0.009341 (rank : 116) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
PHF8_HUMAN
|
||||||
| NC score | 0.008949 (rank : 117) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.008836 (rank : 118) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
TRABD_HUMAN
|
||||||
| NC score | 0.008822 (rank : 119) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4I3, Q96ED8, Q9H7N1, Q9UGX6, Q9UGX7 | Gene names | TRABD | |||
|
Domain Architecture |

|
|||||
| Description | TraB domain-containing protein. | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.008747 (rank : 120) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MICA2_HUMAN
|
||||||
| NC score | 0.008166 (rank : 121) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
CO4A4_HUMAN
|
||||||
| NC score | 0.008057 (rank : 122) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.008033 (rank : 123) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.008015 (rank : 124) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.008009 (rank : 125) | θ value | 0.21417 (rank : 36) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
SI1L1_HUMAN
|
||||||
| NC score | 0.007305 (rank : 126) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.007275 (rank : 127) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
BOC_MOUSE
|
||||||
| NC score | 0.006985 (rank : 128) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6AZB0, Q6KAM5, Q6P5H3, Q7TMJ3, Q8CE73, Q8CE91, Q8R377, Q923W7 | Gene names | Boc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
LRBA_HUMAN
|
||||||
| NC score | 0.006850 (rank : 129) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
ZO2_HUMAN
|
||||||
| NC score | 0.006817 (rank : 130) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.006689 (rank : 131) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
WDHD1_MOUSE
|
||||||
| NC score | 0.006616 (rank : 132) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.006150 (rank : 133) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.005623 (rank : 134) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
AT1A2_HUMAN
|
||||||
| NC score | 0.005311 (rank : 135) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A2_MOUSE
|
||||||
| NC score | 0.005305 (rank : 136) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.005045 (rank : 137) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
SEM6A_HUMAN
|
||||||
| NC score | 0.004815 (rank : 138) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.004785 (rank : 139) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.004471 (rank : 140) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.003592 (rank : 141) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
SLIK3_HUMAN
|
||||||
| NC score | 0.002899 (rank : 142) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94933 | Gene names | SLITRK3, KIAA0848 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLIT and NTRK-like protein 3 precursor. | |||||
|
KI13A_HUMAN
|
||||||
| NC score | 0.002651 (rank : 143) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.002320 (rank : 144) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
SHOC2_HUMAN
|
||||||
| NC score | 0.001779 (rank : 145) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ13, O76063 | Gene names | SHOC2, KIAA0862 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
CP2J5_MOUSE
|
||||||
| NC score | 0.001503 (rank : 146) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54749 | Gene names | Cyp2j5 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2J5 (EC 1.14.14.1) (CYPIIJ5) (Arachidonic acid epoxygenase). | |||||
|
SHOC2_MOUSE
|
||||||
| NC score | 0.001444 (rank : 147) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88520, Q3UJH6, Q8BVL0 | Gene names | Shoc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.000092 (rank : 148) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | -0.000088 (rank : 149) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
NPY4R_HUMAN
|
||||||
| NC score | -0.000562 (rank : 150) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50391, Q13456 | Gene names | PPYR1, NPY4R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropeptide Y receptor type 4 (NPY4-R) (Pancreatic polypeptide receptor 1) (PP1). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | -0.000656 (rank : 151) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | -0.000740 (rank : 152) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||