Please be patient as the page loads
|
DEN1A_HUMAN
|
||||||
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DEN1A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 176 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DEN1A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.967689 (rank : 2) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DEN2A_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 3) | NC score | 0.787729 (rank : 6) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
DEN2A_MOUSE
|
||||||
| θ value | 7.52953e-39 (rank : 4) | NC score | 0.793845 (rank : 5) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
ST5_MOUSE
|
||||||
| θ value | 5.39604e-37 (rank : 5) | NC score | 0.784333 (rank : 8) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
ST5_HUMAN
|
||||||
| θ value | 1.20211e-36 (rank : 6) | NC score | 0.782756 (rank : 10) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
DEN2C_HUMAN
|
||||||
| θ value | 3.4976e-36 (rank : 7) | NC score | 0.786539 (rank : 7) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q68D51, Q5TCX6, Q6P3R3 | Gene names | DENND2C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
DEN2D_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 8) | NC score | 0.812806 (rank : 3) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H6A0, Q5T5V6, Q9BSU0 | Gene names | DENND2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
DEN2C_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 9) | NC score | 0.783755 (rank : 9) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P9P8, Q3T9X2, Q3UV30, Q80V46 | Gene names | Dennd2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
DEN2D_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 10) | NC score | 0.811183 (rank : 4) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91VV4, Q3U028, Q9D831 | Gene names | Dennd2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
RA6I1_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 11) | NC score | 0.601583 (rank : 11) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 12) | NC score | 0.600938 (rank : 12) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
DEN4C_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 13) | NC score | 0.590535 (rank : 14) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VZ89, Q6AI48, Q6ZUB3, Q8NCY7, Q9H6N4, Q9NUT1, Q9NWA5 | Gene names | DENND4C, C9orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 4C. | |||||
|
MYCPP_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 14) | NC score | 0.597887 (rank : 13) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z401, Q14655, Q86T77, Q8IVX2, Q8NB93 | Gene names | MYCPBP, IRLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-myc promoter-binding protein. | |||||
|
MTMRD_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 15) | NC score | 0.435619 (rank : 15) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 16) | NC score | 0.431701 (rank : 16) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 17) | NC score | 0.096389 (rank : 18) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 18) | NC score | 0.085760 (rank : 22) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 19) | NC score | 0.107630 (rank : 17) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 20) | NC score | 0.070169 (rank : 34) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 21) | NC score | 0.079885 (rank : 26) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 22) | NC score | 0.021692 (rank : 145) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
HIC1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 23) | NC score | 0.010875 (rank : 178) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14526 | Gene names | HIC1, ZBTB29 | |||
|
Domain Architecture |
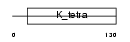
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1) (Zinc finger and BTB domain-containing protein 29). | |||||
|
HIC1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 24) | NC score | 0.011555 (rank : 174) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |
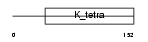
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 25) | NC score | 0.074170 (rank : 31) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 26) | NC score | 0.058981 (rank : 52) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 27) | NC score | 0.067307 (rank : 37) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 28) | NC score | 0.053049 (rank : 67) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 29) | NC score | 0.080968 (rank : 25) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 30) | NC score | 0.043109 (rank : 88) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 31) | NC score | 0.025649 (rank : 126) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 32) | NC score | 0.060860 (rank : 46) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.036206 (rank : 98) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.066395 (rank : 38) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.065558 (rank : 39) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
RFFL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.033869 (rank : 106) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.083903 (rank : 23) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.088782 (rank : 21) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.060086 (rank : 49) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.055679 (rank : 56) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
IRX3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.028632 (rank : 122) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
CO8A1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.031594 (rank : 112) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.074677 (rank : 30) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CO8A1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.032596 (rank : 109) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q00780, Q9D2V4 | Gene names | Col8a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor. | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.051268 (rank : 77) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.079093 (rank : 27) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SH2D3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.034145 (rank : 103) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.052887 (rank : 69) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
USE1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.047472 (rank : 85) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZ43, Q8NCK1, Q9BRT4 | Gene names | USE1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vesicle transport protein USE1 (USE1-like protein) (Putative MAPK- activating protein PM26) (Protein p31). | |||||
|
OR6C4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.000427 (rank : 194) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGE1 | Gene names | OR6C4 | |||
|
Domain Architecture |
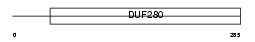
|
|||||
| Description | Olfactory receptor 6C4. | |||||
|
PRAX_MOUSE
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.048973 (rank : 83) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
RBP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.022595 (rank : 140) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.031753 (rank : 110) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.007082 (rank : 183) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.053226 (rank : 66) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ASTL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.030878 (rank : 115) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.092868 (rank : 19) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DHX35_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.019512 (rank : 155) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.048229 (rank : 84) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.031747 (rank : 111) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.036741 (rank : 96) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.090764 (rank : 20) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.040589 (rank : 93) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.020691 (rank : 151) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.047294 (rank : 86) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.081096 (rank : 24) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.065111 (rank : 40) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.029213 (rank : 121) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
RUNX1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.019149 (rank : 158) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.036186 (rank : 99) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.029565 (rank : 119) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.035428 (rank : 102) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.041799 (rank : 89) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.071615 (rank : 33) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.022871 (rank : 139) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.055156 (rank : 60) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CO039_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.053344 (rank : 64) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.024332 (rank : 131) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.024488 (rank : 130) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.036492 (rank : 97) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
NUP98_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.032697 (rank : 108) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.026904 (rank : 125) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.050053 (rank : 80) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.064228 (rank : 42) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.055251 (rank : 59) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
MTF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.004620 (rank : 190) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.075694 (rank : 29) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.038451 (rank : 95) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.040597 (rank : 92) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
RTEL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.024985 (rank : 128) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.009010 (rank : 179) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SRBS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.017773 (rank : 161) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
U689_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.029946 (rank : 117) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D3J8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein UNQ689/PRO1329 homolog precursor. | |||||
|
XKR7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.017465 (rank : 164) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.003505 (rank : 192) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.019210 (rank : 157) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
IRF5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.013866 (rank : 170) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |
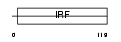
|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
KLF17_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.006734 (rank : 185) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.071801 (rank : 32) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
NELFA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.035693 (rank : 100) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.041726 (rank : 90) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.033624 (rank : 107) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
AF17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.021076 (rank : 150) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
ASPC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.024674 (rank : 129) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VBT9, Q3UKD1, Q6V7K5, Q9CT64 | Gene names | Aspscr1, Tug | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tether containing UBX domain for GLUT4 (Alveolar soft part sarcoma chromosome region candidate 1 homolog). | |||||
|
COCA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.011640 (rank : 173) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.033880 (rank : 105) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
PCD15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.006199 (rank : 186) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.017515 (rank : 163) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RU1C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.051729 (rank : 75) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09234 | Gene names | SNRPC | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.062635 (rank : 44) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.022250 (rank : 143) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZN253_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.002703 (rank : 193) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75346 | Gene names | ZNF253, BMZF1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 253 (Bone marrow zinc finger 1) (BMZF-1). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.023003 (rank : 137) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.004393 (rank : 191) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.031169 (rank : 113) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.023713 (rank : 133) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
GRIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.006998 (rank : 184) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
IF4G2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.025191 (rank : 127) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62448, P97867, Q921U3 | Gene names | Eif4g2, Nat1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Novel APOBEC-1 target 1) (Translation repressor NAT1). | |||||
|
MINP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.023827 (rank : 132) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2L6, Q8VDR0, Q9JJD5 | Gene names | Minpp1, Mipp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple inositol polyphosphate phosphatase 1 precursor (EC 3.1.3.62) (Inositol (1,3,4,5)-tetrakisphosphate 3-phosphatase) (Ins(1,3,4,5)P(4) 3-phosphatase). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.067601 (rank : 36) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.019791 (rank : 154) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.034084 (rank : 104) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PHLP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.021104 (rank : 149) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13371, Q96AF1, Q9UEW7, Q9UFL0 | Gene names | PDCL | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.030190 (rank : 116) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.011011 (rank : 176) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.010927 (rank : 177) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.023093 (rank : 136) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.064033 (rank : 43) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN703_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.029887 (rank : 118) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H7S9, Q5XG76 | Gene names | ZNF703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 703. | |||||
|
ATG4D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.008585 (rank : 180) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86TL0 | Gene names | ATG4D, APG4D, AUTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine protease ATG4D (EC 3.4.22.-) (Autophagy-related protein 4 homolog D) (Autophagin-4) (Autophagy-related cysteine endopeptidase 4) (AUT-like 4 cysteine endopeptidase). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.059300 (rank : 51) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.022885 (rank : 138) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.051319 (rank : 76) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.051757 (rank : 74) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.018099 (rank : 160) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.050028 (rank : 81) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
FIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.020688 (rank : 152) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
GGN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.057613 (rank : 53) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GP179_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.022077 (rank : 144) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.021173 (rank : 148) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HPS4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.023354 (rank : 135) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
M3K14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | -0.001216 (rank : 195) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUL6 | Gene names | Map3k14, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK). | |||||
|
M3K5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | -0.003181 (rank : 196) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35099 | Gene names | Map3k5, Ask1, Mekk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 5 (EC 2.7.11.25) (MAPK/ERK kinase kinase 5) (MEK kinase 5) (MEKK 5) (Apoptosis signal- regulating kinase 1) (ASK-1). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.021296 (rank : 147) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.059316 (rank : 50) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.017406 (rank : 165) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.040038 (rank : 94) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
RU1C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.045645 (rank : 87) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62241 | Gene names | Snrp1c, Snrpc | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.030995 (rank : 114) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
T240L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.019116 (rank : 159) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.023545 (rank : 134) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.019944 (rank : 153) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.019217 (rank : 156) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
USBP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.012616 (rank : 172) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N6Y0, Q8NBX7, Q96KH3, Q9BYI8 | Gene names | USHBP1, AIEBP, MCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1) (MCC-2) (AIE-75 binding protein). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.060265 (rank : 48) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
AMBN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.016338 (rank : 167) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NP70, Q9H2X1, Q9H4L1 | Gene names | AMBN | |||
|
Domain Architecture |
|
|||||
| Description | Ameloblastin precursor. | |||||
|
BBX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.011199 (rank : 175) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.035671 (rank : 101) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.015001 (rank : 168) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.012648 (rank : 171) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.028587 (rank : 123) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
IF4G2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.021488 (rank : 146) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78344, O60877, P78404, Q53EU1, Q58EZ2, Q8NI71, Q96C16 | Gene names | EIF4G2, DAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Death-associated protein 5) (DAP-5). | |||||
|
LITAF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.014275 (rank : 169) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLJ0, Q9EQI0 | Gene names | Litaf, N4wbp3, Tbx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipopolysaccharide-induced tumor necrosis factor-alpha factor homolog (LPS-induced TNF-alpha factor homolog) (LITAF-like protein) (NEDD4 WW domain-binding protein 3) (Estrogen-enhanced transcript protein) (mEET). | |||||
|
NUMBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.016988 (rank : 166) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.064588 (rank : 41) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.027731 (rank : 124) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
RPO3F_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.022458 (rank : 142) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1D9, O15319 | Gene names | POLR3F, RPC39 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-directed RNA polymerase III subunit F (EC 2.7.7.6) (DNA-directed RNA polymerase III 39 kDa polypeptide) (RNA polymerase III C39 subunit). | |||||
|
RPO3F_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.022461 (rank : 141) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921X6, Q544K6 | Gene names | Polr3f | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III subunit F (EC 2.7.7.6) (DNA-directed RNA polymerase III 39 kDa polypeptide) (RNA polymerase III C39 subunit). | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.007244 (rank : 182) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SGIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.029346 (rank : 120) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.005043 (rank : 189) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
STAU1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.007487 (rank : 181) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.017703 (rank : 162) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
TEAD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.005253 (rank : 188) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62296, P70282, Q61174, Q61175, Q62298 | Gene names | Tead4, Tcf13r1, Tef3, Tefr1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcriptional enhancer factor TEF-3 (TEA domain family member 4) (TEAD-4) (ETF-related factor 2) (ETFR-2) (TEF-1-related factor 1) (TEF-1-related factor FR-19) (RTEF-1). | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.005734 (rank : 187) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.041175 (rank : 91) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.050519 (rank : 79) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.052240 (rank : 72) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.055388 (rank : 58) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.050001 (rank : 82) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.061823 (rank : 45) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.060798 (rank : 47) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.051041 (rank : 78) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.067786 (rank : 35) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.077787 (rank : 28) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.056173 (rank : 55) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.051929 (rank : 73) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RERE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.052520 (rank : 71) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.053018 (rank : 68) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.053838 (rank : 62) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.057195 (rank : 54) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.054042 (rank : 61) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.052831 (rank : 70) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.053639 (rank : 63) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.055663 (rank : 57) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.053243 (rank : 65) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
DEN1A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 176 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DEN1A_MOUSE
|
||||||
| NC score | 0.967689 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DEN2D_HUMAN
|
||||||
| NC score | 0.812806 (rank : 3) | θ value | 2.96089e-35 (rank : 8) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H6A0, Q5T5V6, Q9BSU0 | Gene names | DENND2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
DEN2D_MOUSE
|
||||||
| NC score | 0.811183 (rank : 4) | θ value | 1.46948e-34 (rank : 10) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91VV4, Q3U028, Q9D831 | Gene names | Dennd2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
DEN2A_MOUSE
|
||||||
| NC score | 0.793845 (rank : 5) | θ value | 7.52953e-39 (rank : 4) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
DEN2A_HUMAN
|
||||||
| NC score | 0.787729 (rank : 6) | θ value | 8.89434e-40 (rank : 3) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
DEN2C_HUMAN
|
||||||
| NC score | 0.786539 (rank : 7) | θ value | 3.4976e-36 (rank : 7) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q68D51, Q5TCX6, Q6P3R3 | Gene names | DENND2C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
ST5_MOUSE
|
||||||
| NC score | 0.784333 (rank : 8) | θ value | 5.39604e-37 (rank : 5) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
DEN2C_MOUSE
|
||||||
| NC score | 0.783755 (rank : 9) | θ value | 1.12514e-34 (rank : 9) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P9P8, Q3T9X2, Q3UV30, Q80V46 | Gene names | Dennd2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
ST5_HUMAN
|
||||||
| NC score | 0.782756 (rank : 10) | θ value | 1.20211e-36 (rank : 6) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
RA6I1_HUMAN
|
||||||
| NC score | 0.601583 (rank : 11) | θ value | 2.13179e-17 (rank : 11) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.600938 (rank : 12) | θ value | 4.74913e-17 (rank : 12) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
MYCPP_HUMAN
|
||||||
| NC score | 0.597887 (rank : 13) | θ value | 2.20605e-14 (rank : 14) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z401, Q14655, Q86T77, Q8IVX2, Q8NB93 | Gene names | MYCPBP, IRLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-myc promoter-binding protein. | |||||
|
DEN4C_HUMAN
|
||||||
| NC score | 0.590535 (rank : 14) | θ value | 9.90251e-15 (rank : 13) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VZ89, Q6AI48, Q6ZUB3, Q8NCY7, Q9H6N4, Q9NUT1, Q9NWA5 | Gene names | DENND4C, C9orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 4C. | |||||
|
MTMRD_HUMAN
|
||||||
| NC score | 0.435619 (rank : 15) | θ value | 3.18553e-13 (rank : 15) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.431701 (rank : 16) | θ value | 6.00763e-12 (rank : 16) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.107630 (rank : 17) | θ value | 0.00665767 (rank : 19) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.096389 (rank : 18) | θ value | 4.1701e-05 (rank : 17) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.092868 (rank : 19) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.090764 (rank : 20) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.088782 (rank : 21) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.085760 (rank : 22) | θ value | 0.00228821 (rank : 18) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.083903 (rank : 23) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.081096 (rank : 24) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.080968 (rank : 25) | θ value | 0.0431538 (rank : 29) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.079885 (rank : 26) | θ value | 0.0193708 (rank : 21) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.079093 (rank : 27) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.077787 (rank : 28) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.075694 (rank : 29) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.074677 (rank : 30) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.074170 (rank : 31) | θ value | 0.0252991 (rank : 25) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.071801 (rank : 32) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.071615 (rank : 33) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.070169 (rank : 34) | θ value | 0.00665767 (rank : 20) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.067786 (rank : 35) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.067601 (rank : 36) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.067307 (rank : 37) | θ value | 0.0330416 (rank : 27) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.066395 (rank : 38) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.065558 (rank : 39) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.065111 (rank : 40) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.064588 (rank : 41) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.064228 (rank : 42) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.064033 (rank : 43) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.062635 (rank : 44) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.061823 (rank : 45) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.060860 (rank : 46) | θ value | 0.0736092 (rank : 32) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.060798 (rank : 47) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.060265 (rank : 48) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.060086 (rank : 49) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.059316 (rank : 50) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.059300 (rank : 51) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.058981 (rank : 52) | θ value | 0.0330416 (rank : 26) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.057613 (rank : 53) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.057195 (rank : 54) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.056173 (rank : 55) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.055679 (rank : 56) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.055663 (rank : 57) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.055388 (rank : 58) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.055251 (rank : 59) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.055156 (rank : 60) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.054042 (rank : 61) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.053838 (rank : 62) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.053639 (rank : 63) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.053344 (rank : 64) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.053243 (rank : 65) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.053226 (rank : 66) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.053049 (rank : 67) | θ value | 0.0431538 (rank : 28) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.053018 (rank : 68) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.052887 (rank : 69) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.052831 (rank : 70) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.052520 (rank : 71) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.052240 (rank : 72) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.051929 (rank : 73) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.051757 (rank : 74) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
RU1C_HUMAN
|
||||||
| NC score | 0.051729 (rank : 75) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09234 | Gene names | SNRPC | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.051319 (rank : 76) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.051268 (rank : 77) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.051041 (rank : 78) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.050519 (rank : 79) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.050053 (rank : 80) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.050028 (rank : 81) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.050001 (rank : 82) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PRAX_MOUSE
|
||||||
| NC score | 0.048973 (rank : 83) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.048229 (rank : 84) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
USE1_HUMAN
|
||||||
| NC score | 0.047472 (rank : 85) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZ43, Q8NCK1, Q9BRT4 | Gene names | USE1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vesicle transport protein USE1 (USE1-like protein) (Putative MAPK- activating protein PM26) (Protein p31). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.047294 (rank : 86) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
RU1C_MOUSE
|
||||||
| NC score | 0.045645 (rank : 87) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62241 | Gene names | Snrp1c, Snrpc | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.043109 (rank : 88) | θ value | 0.0563607 (rank : 30) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.041799 (rank : 89) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.041726 (rank : 90) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.041175 (rank : 91) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.040597 (rank : 92) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.040589 (rank : 93) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.040038 (rank : 94) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.038451 (rank : 95) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.036741 (rank : 96) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.036492 (rank : 97) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.036206 (rank : 98) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.036186 (rank : 99) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
NELFA_HUMAN
|
||||||
| NC score | 0.035693 (rank : 100) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.035671 (rank : 101) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.035428 (rank : 102) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
SH2D3_HUMAN
|
||||||
| NC score | 0.034145 (rank : 103) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.034084 (rank : 104) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.033880 (rank : 105) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
RFFL_HUMAN
|
||||||
| NC score | 0.033869 (rank : 106) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.033624 (rank : 107) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
NUP98_HUMAN
|
||||||
| NC score | 0.032697 (rank : 108) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
CO8A1_MOUSE
|
||||||
| NC score | 0.032596 (rank : 109) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q00780, Q9D2V4 | Gene names | Col8a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor. | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.031753 (rank : 110) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.031747 (rank : 111) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
CO8A1_HUMAN
|
||||||
| NC score | 0.031594 (rank : 112) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.031169 (rank : 113) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.030995 (rank : 114) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ASTL_HUMAN
|
||||||
| NC score | 0.030878 (rank : 115) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.030190 (rank : 116) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
U689_MOUSE
|
||||||
| NC score | 0.029946 (rank : 117) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D3J8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein UNQ689/PRO1329 homolog precursor. | |||||
|
ZN703_HUMAN
|
||||||
| NC score | 0.029887 (rank : 118) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H7S9, Q5XG76 | Gene names | ZNF703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 703. | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.029565 (rank : 119) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
SGIP1_MOUSE
|
||||||
| NC score | 0.029346 (rank : 120) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.029213 (rank : 121) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
IRX3_MOUSE
|
||||||
| NC score | 0.028632 (rank : 122) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.028587 (rank : 123) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.027731 (rank : 124) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.026904 (rank : 125) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.025649 (rank : 126) | θ value | 0.0736092 (rank : 31) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
IF4G2_MOUSE
|
||||||
| NC score | 0.025191 (rank : 127) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62448, P97867, Q921U3 | Gene names | Eif4g2, Nat1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Novel APOBEC-1 target 1) (Translation repressor NAT1). | |||||
|
RTEL1_HUMAN
|
||||||
| NC score | 0.024985 (rank : 128) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
ASPC1_MOUSE
|
||||||
| NC score | 0.024674 (rank : 129) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VBT9, Q3UKD1, Q6V7K5, Q9CT64 | Gene names | Aspscr1, Tug | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tether containing UBX domain for GLUT4 (Alveolar soft part sarcoma chromosome region candidate 1 homolog). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.024488 (rank : 130) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.024332 (rank : 131) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
MINP1_MOUSE
|
||||||
| NC score | 0.023827 (rank : 132) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2L6, Q8VDR0, Q9JJD5 | Gene names | Minpp1, Mipp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple inositol polyphosphate phosphatase 1 precursor (EC 3.1.3.62) (Inositol (1,3,4,5)-tetrakisphosphate 3-phosphatase) (Ins(1,3,4,5)P(4) 3-phosphatase). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.023713 (rank : 133) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.023545 (rank : 134) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
HPS4_MOUSE
|
||||||
| NC score | 0.023354 (rank : 135) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.023093 (rank : 136) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.023003 (rank : 137) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.022885 (rank : 138) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.022871 (rank : 139) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
RBP2_HUMAN
|
||||||
| NC score | 0.022595 (rank : 140) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
RPO3F_MOUSE
|
||||||
| NC score | 0.022461 (rank : 141) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921X6, Q544K6 | Gene names | Polr3f | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III subunit F (EC 2.7.7.6) (DNA-directed RNA polymerase III 39 kDa polypeptide) (RNA polymerase III C39 subunit). | |||||
|
RPO3F_HUMAN
|
||||||
| NC score | 0.022458 (rank : 142) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1D9, O15319 | Gene names | POLR3F, RPC39 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-directed RNA polymerase III subunit F (EC 2.7.7.6) (DNA-directed RNA polymerase III 39 kDa polypeptide) (RNA polymerase III C39 subunit). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.022250 (rank : 143) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.022077 (rank : 144) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.021692 (rank : 145) | θ value | 0.0193708 (rank : 22) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
IF4G2_HUMAN
|
||||||
| NC score | 0.021488 (rank : 146) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78344, O60877, P78404, Q53EU1, Q58EZ2, Q8NI71, Q96C16 | Gene names | EIF4G2, DAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Death-associated protein 5) (DAP-5). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.021296 (rank : 147) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.021173 (rank : 148) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
PHLP_HUMAN
|
||||||
| NC score | 0.021104 (rank : 149) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13371, Q96AF1, Q9UEW7, Q9UFL0 | Gene names | PDCL | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.021076 (rank : 150) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.020691 (rank : 151) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
FIP1_HUMAN
|
||||||
| NC score | 0.020688 (rank : 152) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.019944 (rank : 153) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.019791 (rank : 154) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
DHX35_HUMAN
|
||||||
| NC score | 0.019512 (rank : 155) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.019217 (rank : 156) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.019210 (rank : 157) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
RUNX1_HUMAN
|
||||||
| NC score | 0.019149 (rank : 158) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
T240L_HUMAN
|
||||||
| NC score | 0.019116 (rank : 159) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.018099 (rank : 160) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
SRBS1_HUMAN
|
||||||
| NC score | 0.017773 (rank : 161) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.017703 (rank : 162) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.017515 (rank : 163) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
XKR7_MOUSE
|
||||||
| NC score | 0.017465 (rank : 164) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.017406 (rank : 165) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
NUMBL_MOUSE
|
||||||
| NC score | 0.016988 (rank : 166) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
AMBN_HUMAN
|
||||||
| NC score | 0.016338 (rank : 167) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NP70, Q9H2X1, Q9H4L1 | Gene names | AMBN | |||
|
Domain Architecture |

|
|||||
| Description | Ameloblastin precursor. | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.015001 (rank : 168) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
LITAF_MOUSE
|
||||||
| NC score | 0.014275 (rank : 169) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLJ0, Q9EQI0 | Gene names | Litaf, N4wbp3, Tbx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipopolysaccharide-induced tumor necrosis factor-alpha factor homolog (LPS-induced TNF-alpha factor homolog) (LITAF-like protein) (NEDD4 WW domain-binding protein 3) (Estrogen-enhanced transcript protein) (mEET). | |||||
|
IRF5_MOUSE
|
||||||
| NC score | 0.013866 (rank : 170) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.012648 (rank : 171) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
USBP1_HUMAN
|
||||||
| NC score | 0.012616 (rank : 172) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N6Y0, Q8NBX7, Q96KH3, Q9BYI8 | Gene names | USHBP1, AIEBP, MCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1) (MCC-2) (AIE-75 binding protein). | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.011640 (rank : 173) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
HIC1_MOUSE
|
||||||
| NC score | 0.011555 (rank : 174) | θ value | 0.0252991 (rank : 24) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
BBX_MOUSE
|
||||||
| NC score | 0.011199 (rank : 175) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.011011 (rank : 176) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.010927 (rank : 177) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
HIC1_HUMAN
|
||||||
| NC score | 0.010875 (rank : 178) | θ value | 0.0252991 (rank : 23) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14526 | Gene names | HIC1, ZBTB29 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1) (Zinc finger and BTB domain-containing protein 29). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.009010 (rank : 179) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ATG4D_HUMAN
|
||||||
| NC score | 0.008585 (rank : 180) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86TL0 | Gene names | ATG4D, APG4D, AUTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine protease ATG4D (EC 3.4.22.-) (Autophagy-related protein 4 homolog D) (Autophagin-4) (Autophagy-related cysteine endopeptidase 4) (AUT-like 4 cysteine endopeptidase). | |||||
|
STAU1_HUMAN
|
||||||
| NC score | 0.007487 (rank : 181) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.007244 (rank : 182) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.007082 (rank : 183) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
GRIP1_MOUSE
|
||||||
| NC score | 0.006998 (rank : 184) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
KLF17_HUMAN
|
||||||
| NC score | 0.006734 (rank : 185) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
PCD15_HUMAN
|
||||||
| NC score | 0.006199 (rank : 186) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.005734 (rank : 187) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TEAD4_MOUSE
|
||||||
| NC score | 0.005253 (rank : 188) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62296, P70282, Q61174, Q61175, Q62298 | Gene names | Tead4, Tcf13r1, Tef3, Tefr1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional enhancer factor TEF-3 (TEA domain family member 4) (TEAD-4) (ETF-related factor 2) (ETFR-2) (TEF-1-related factor 1) (TEF-1-related factor FR-19) (RTEF-1). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.005043 (rank : 189) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
MTF1_HUMAN
|
||||||
| NC score | 0.004620 (rank : 190) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.004393 (rank : 191) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | 0.003505 (rank : 192) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ZN253_HUMAN
|
||||||
| NC score | 0.002703 (rank : 193) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75346 | Gene names | ZNF253, BMZF1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 253 (Bone marrow zinc finger 1) (BMZF-1). | |||||
|
OR6C4_HUMAN
|
||||||
| NC score | 0.000427 (rank : 194) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGE1 | Gene names | OR6C4 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 6C4. | |||||
|
M3K14_MOUSE
|
||||||
| NC score | -0.001216 (rank : 195) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUL6 | Gene names | Map3k14, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK). | |||||
|
M3K5_MOUSE
|
||||||
| NC score | -0.003181 (rank : 196) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 176 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35099 | Gene names | Map3k5, Ask1, Mekk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 5 (EC 2.7.11.25) (MAPK/ERK kinase kinase 5) (MEK kinase 5) (MEKK 5) (Apoptosis signal- regulating kinase 1) (ASK-1). | |||||