Please be patient as the page loads
|
NUP98_HUMAN
|
||||||
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NUP98_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
TROP_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 2) | NC score | 0.115221 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 3) | NC score | 0.108063 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 4) | NC score | 0.138568 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 5) | NC score | 0.143372 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
CK024_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.111925 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.079043 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CPSF7_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.114785 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
CPSF7_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.114842 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
PO121_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.129243 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.065537 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.059745 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
PHXR5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.175740 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.059091 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
NUP62_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.103367 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
RNPL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.043271 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAU8, Q96AC9, Q9H033, Q9NVD0 | Gene names | RNPEPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl aminopeptidase-like 1 (EC 3.4.11.-) (RNPEP-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.034159 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.045633 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PUM1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.057844 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U78, Q80X96, Q80YU8, Q8BPV7, Q9EPU6 | Gene names | Pum1, Kiaa0099 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 1. | |||||
|
MCM3A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.058919 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.037003 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.032634 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
PUM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.052720 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14671, Q5VXY7, Q9HAN1 | Gene names | PUM1, KIAA0099, PUMH1 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 1 (Pumilio-1) (HsPUM). | |||||
|
DEN1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.032697 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
EYA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.028426 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
HAIR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.031205 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.020112 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PAPOA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.038919 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.012237 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
K0586_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.044993 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
WAPL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.043061 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z5K2, Q5VSK5, Q8IX10, Q92549 | Gene names | WAPAL, FOE, KIAA0261, WAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Friend of EBNA2 protein). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.015425 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ESCO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.030426 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z69, Q8BQI2, Q8BR47, Q922F5 | Gene names | Esco1, Kiaa1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1). | |||||
|
MTFR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.036257 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
|
MTP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.033292 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55157 | Gene names | MTTP, MTP | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal triglyceride transfer protein large subunit precursor. | |||||
|
NUP54_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.046816 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z3B4, Q96EA7, Q9NVL5, Q9P0I1 | Gene names | NUP54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin p54 (54 kDa nucleoporin). | |||||
|
SALL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.004783 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.025152 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
VRK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.022089 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IV63, Q9P2V8 | Gene names | VRK3 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase VRK3 (EC 2.7.11.1) (Vaccinia-related kinase 3). | |||||
|
ANPRA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.006717 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
BUB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.021257 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
CSTF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.016112 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.032216 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
NPHP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.022382 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15259, O14837 | Gene names | NPHP1, NPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1 (Juvenile nephronophthisis 1 protein). | |||||
|
NUP62_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.087251 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.090371 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
CASPA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.018408 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.018869 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.014174 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
FOXO3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.014247 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43524, O15171, Q5T2I7, Q9BZ04 | Gene names | FOXO3A, FKHRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O3A (Forkhead in rhabdomyosarcoma-like 1) (AF6q21 protein). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.009457 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
NUP53_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.032590 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
PTCD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.027815 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75127, Q66K60, Q9UDV2 | Gene names | PTCD1, KIAA0632 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pentatricopeptide repeat protein 1. | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.018085 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
BRCA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.015428 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
CLCN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.011515 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35523 | Gene names | CLCN1, CLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein, skeletal muscle (Chloride channel protein 1) (ClC-1). | |||||
|
CX6A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.027147 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12074, O43714, Q32Q37 | Gene names | COX6A1, COX6AL | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome c oxidase polypeptide VIa-liver, mitochondrial precursor (EC 1.9.3.1). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.014078 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
LIS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.007420 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63005, O35592, P43035, P81692, Q9R2A6 | Gene names | Pafah1b1, Lis-1, Lis1, Pafaha | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-activating factor acetylhydrolase IB subunit alpha (PAF acetylhydrolase 45 kDa subunit) (PAF-AH 45 kDa subunit) (PAF-AH alpha) (PAFAH alpha) (Lissencephaly-1 protein) (LIS-1). | |||||
|
MUCEN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.039980 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
PAPOA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.026504 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
PCDB4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.002980 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5E5 | Gene names | PCDHB4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 4 precursor (PCDH-beta4). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.018069 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.006688 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.021984 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.006224 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CN135_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.024354 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q63HM2, Q9BQG8, Q9H9F2 | Gene names | C14orf135, FBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf135 precursor (Hepatitis C virus F protein-binding protein 2) (HCV F protein-binding protein 2). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.008254 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.023194 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
FBSH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.021042 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HAH7, Q96CI9, Q9H9X4 | Gene names | FBS1, FBS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable fibrosin-1 long transcript protein. | |||||
|
GATA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.010537 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
K0562_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.018095 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80V31, Q6ZQ95 | Gene names | Kiaa0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.009596 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
LIS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.006681 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43034, Q8WZ88, Q8WZ89 | Gene names | PAFAH1B1, LIS1, MDCR, PAFAHA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet-activating factor acetylhydrolase IB subunit alpha (PAF acetylhydrolase 45 kDa subunit) (PAF-AH 45 kDa subunit) (PAF-AH alpha) (PAFAH alpha) (Lissencephaly-1 protein) (LIS-1). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.017899 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
PML_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.014017 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60953 | Gene names | Pml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable transcription factor PML. | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.005322 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.030931 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.016254 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TSKS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.016190 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJT2, Q8WXJ0 | Gene names | STK22S1, TSKS, TSKS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
WAPL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.032894 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.055663 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CO002_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.050548 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
LORI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.097333 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LORI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.054084 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
SON_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.051503 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
NUP98_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
PHXR5_MOUSE
|
||||||
| NC score | 0.175740 (rank : 2) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.143372 (rank : 3) | θ value | 0.0148317 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.138568 (rank : 4) | θ value | 0.0113563 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.129243 (rank : 5) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
TROP_HUMAN
|
||||||
| NC score | 0.115221 (rank : 6) | θ value | 2.88788e-06 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
CPSF7_MOUSE
|
||||||
| NC score | 0.114842 (rank : 7) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
CPSF7_HUMAN
|
||||||
| NC score | 0.114785 (rank : 8) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.111925 (rank : 9) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.108063 (rank : 10) | θ value | 5.44631e-05 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
NUP62_HUMAN
|
||||||
| NC score | 0.103367 (rank : 11) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.097333 (rank : 12) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.090371 (rank : 13) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
NUP62_MOUSE
|
||||||
| NC score | 0.087251 (rank : 14) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.079043 (rank : 15) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.065537 (rank : 16) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.059745 (rank : 17) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.059091 (rank : 18) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
MCM3A_MOUSE
|
||||||
| NC score | 0.058919 (rank : 19) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
PUM1_MOUSE
|
||||||
| NC score | 0.057844 (rank : 20) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U78, Q80X96, Q80YU8, Q8BPV7, Q9EPU6 | Gene names | Pum1, Kiaa0099 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 1. | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.055663 (rank : 21) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.054084 (rank : 22) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
PUM1_HUMAN
|
||||||
| NC score | 0.052720 (rank : 23) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14671, Q5VXY7, Q9HAN1 | Gene names | PUM1, KIAA0099, PUMH1 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 1 (Pumilio-1) (HsPUM). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.051503 (rank : 24) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.050548 (rank : 25) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
NUP54_HUMAN
|
||||||
| NC score | 0.046816 (rank : 26) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z3B4, Q96EA7, Q9NVL5, Q9P0I1 | Gene names | NUP54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin p54 (54 kDa nucleoporin). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.045633 (rank : 27) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
K0586_HUMAN
|
||||||
| NC score | 0.044993 (rank : 28) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
RNPL1_HUMAN
|
||||||
| NC score | 0.043271 (rank : 29) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAU8, Q96AC9, Q9H033, Q9NVD0 | Gene names | RNPEPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl aminopeptidase-like 1 (EC 3.4.11.-) (RNPEP-like protein). | |||||
|
WAPL_HUMAN
|
||||||
| NC score | 0.043061 (rank : 30) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z5K2, Q5VSK5, Q8IX10, Q92549 | Gene names | WAPAL, FOE, KIAA0261, WAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Friend of EBNA2 protein). | |||||
|
MUCEN_MOUSE
|
||||||
| NC score | 0.039980 (rank : 31) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
PAPOA_MOUSE
|
||||||
| NC score | 0.038919 (rank : 32) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.037003 (rank : 33) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MTFR1_MOUSE
|
||||||
| NC score | 0.036257 (rank : 34) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.034159 (rank : 35) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MTP_HUMAN
|
||||||
| NC score | 0.033292 (rank : 36) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55157 | Gene names | MTTP, MTP | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal triglyceride transfer protein large subunit precursor. | |||||
|
WAPL_MOUSE
|
||||||
| NC score | 0.032894 (rank : 37) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
DEN1A_HUMAN
|
||||||
| NC score | 0.032697 (rank : 38) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.032634 (rank : 39) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
NUP53_HUMAN
|
||||||
| NC score | 0.032590 (rank : 40) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.032216 (rank : 41) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
HAIR_MOUSE
|
||||||
| NC score | 0.031205 (rank : 42) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.030931 (rank : 43) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
ESCO1_MOUSE
|
||||||
| NC score | 0.030426 (rank : 44) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z69, Q8BQI2, Q8BR47, Q922F5 | Gene names | Esco1, Kiaa1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1). | |||||
|
EYA1_MOUSE
|
||||||
| NC score | 0.028426 (rank : 45) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
PTCD1_HUMAN
|
||||||
| NC score | 0.027815 (rank : 46) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75127, Q66K60, Q9UDV2 | Gene names | PTCD1, KIAA0632 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pentatricopeptide repeat protein 1. | |||||
|
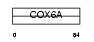
CX6A1_HUMAN
|
||||||
| NC score | 0.027147 (rank : 47) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12074, O43714, Q32Q37 | Gene names | COX6A1, COX6AL | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome c oxidase polypeptide VIa-liver, mitochondrial precursor (EC 1.9.3.1). | |||||
|
PAPOA_HUMAN
|
||||||
| NC score | 0.026504 (rank : 48) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.025152 (rank : 49) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
CN135_HUMAN
|
||||||
| NC score | 0.024354 (rank : 50) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q63HM2, Q9BQG8, Q9H9F2 | Gene names | C14orf135, FBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf135 precursor (Hepatitis C virus F protein-binding protein 2) (HCV F protein-binding protein 2). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.023194 (rank : 51) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
NPHP1_HUMAN
|
||||||
| NC score | 0.022382 (rank : 52) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15259, O14837 | Gene names | NPHP1, NPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1 (Juvenile nephronophthisis 1 protein). | |||||
|
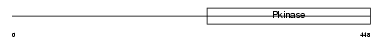
VRK3_HUMAN
|
||||||
| NC score | 0.022089 (rank : 53) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IV63, Q9P2V8 | Gene names | VRK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase VRK3 (EC 2.7.11.1) (Vaccinia-related kinase 3). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.021984 (rank : 54) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
BUB1_HUMAN
|
||||||
| NC score | 0.021257 (rank : 55) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
FBSH_HUMAN
|
||||||
| NC score | 0.021042 (rank : 56) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HAH7, Q96CI9, Q9H9X4 | Gene names | FBS1, FBS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable fibrosin-1 long transcript protein. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.020112 (rank : 57) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.018869 (rank : 58) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
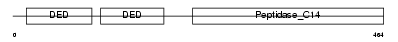
CASPA_HUMAN
|
||||||
| NC score | 0.018408 (rank : 59) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
K0562_MOUSE
|
||||||
| NC score | 0.018095 (rank : 60) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80V31, Q6ZQ95 | Gene names | Kiaa0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.018085 (rank : 61) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.018069 (rank : 62) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.017899 (rank : 63) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.016254 (rank : 64) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TSKS_HUMAN
|
||||||
| NC score | 0.016190 (rank : 65) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJT2, Q8WXJ0 | Gene names | STK22S1, TSKS, TSKS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
CSTF2_MOUSE
|
||||||
| NC score | 0.016112 (rank : 66) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
BRCA1_HUMAN
|
||||||
| NC score | 0.015428 (rank : 67) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.015425 (rank : 68) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
FOXO3_HUMAN
|
||||||
| NC score | 0.014247 (rank : 69) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43524, O15171, Q5T2I7, Q9BZ04 | Gene names | FOXO3A, FKHRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O3A (Forkhead in rhabdomyosarcoma-like 1) (AF6q21 protein). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.014174 (rank : 70) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.014078 (rank : 71) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
PML_MOUSE
|
||||||
| NC score | 0.014017 (rank : 72) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60953 | Gene names | Pml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable transcription factor PML. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.012237 (rank : 73) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CLCN1_HUMAN
|
||||||
| NC score | 0.011515 (rank : 74) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35523 | Gene names | CLCN1, CLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein, skeletal muscle (Chloride channel protein 1) (ClC-1). | |||||
|
GATA6_HUMAN
|
||||||
| NC score | 0.010537 (rank : 75) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.009596 (rank : 76) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.009457 (rank : 77) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.008254 (rank : 78) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
LIS1_MOUSE
|
||||||
| NC score | 0.007420 (rank : 79) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63005, O35592, P43035, P81692, Q9R2A6 | Gene names | Pafah1b1, Lis-1, Lis1, Pafaha | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-activating factor acetylhydrolase IB subunit alpha (PAF acetylhydrolase 45 kDa subunit) (PAF-AH 45 kDa subunit) (PAF-AH alpha) (PAFAH alpha) (Lissencephaly-1 protein) (LIS-1). | |||||
|
ANPRA_MOUSE
|
||||||
| NC score | 0.006717 (rank : 80) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.006688 (rank : 81) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
LIS1_HUMAN
|
||||||
| NC score | 0.006681 (rank : 82) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43034, Q8WZ88, Q8WZ89 | Gene names | PAFAH1B1, LIS1, MDCR, PAFAHA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet-activating factor acetylhydrolase IB subunit alpha (PAF acetylhydrolase 45 kDa subunit) (PAF-AH 45 kDa subunit) (PAF-AH alpha) (PAFAH alpha) (Lissencephaly-1 protein) (LIS-1). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.006224 (rank : 83) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.005322 (rank : 84) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SALL1_HUMAN
|
||||||
| NC score | 0.004783 (rank : 85) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
PCDB4_HUMAN
|
||||||
| NC score | 0.002980 (rank : 86) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5E5 | Gene names | PCDHB4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 4 precursor (PCDH-beta4). | |||||