Please be patient as the page loads
|
PAPOA_MOUSE
|
||||||
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PAPOA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987663 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
PAPOA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
PAPOB_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.981419 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRJ5, Q8NE14 | Gene names | PAPOLB, PAPT | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase beta (EC 2.7.7.19) (PAP beta) (Polynucleotide adenylyltransferase beta) (Testis-specific poly(A) polymerase). | |||||
|
PAPOB_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.983091 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVP6, Q9R1R3 | Gene names | Papolb, Papt, Tpap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase beta (EC 2.7.7.19) (PAP beta) (Polynucleotide adenylyltransferase beta) (Testis-specific poly(A) polymerase). | |||||
|
PAPOG_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.972305 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BWT3, Q969N1, Q9H8L2, Q9HAD0 | Gene names | PAPOLG, PAP2, PAPG | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme) (Neo- poly(A) polymerase) (Neo-PAP). | |||||
|
PAPOG_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.967205 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PCL9, Q8BZC9 | Gene names | Papolg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme). | |||||
|
CK024_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 7) | NC score | 0.117576 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.088502 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.053973 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 10) | NC score | 0.085455 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.036085 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
GA2L3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 12) | NC score | 0.056873 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.039931 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 14) | NC score | 0.082975 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.057676 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.061470 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.032180 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.057149 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
CD34_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.064268 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64314, Q62550, Q62551 | Gene names | Cd34 | |||
|
Domain Architecture |
|
|||||
| Description | Hematopoietic progenitor cell antigen CD34 precursor. | |||||
|
DAF1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.021779 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.027969 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
ZBT38_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.004532 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
LRC27_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.026618 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80YS5, Q9D6Z2 | Gene names | Lrrc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.069232 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
RXRA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.008343 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
SYNP2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.034827 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.045859 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.039073 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PALM_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.033947 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
TAF9B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.028783 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HBM6, Q9Y2S3 | Gene names | TAF9B, TAF9L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 9B (Transcription initiation factor TFIID subunit 9-like protein) (Transcription- associated factor TAFII31L) (Neuronal cell death-related protein 7) (DN-7). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.067093 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.033474 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.023457 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.013671 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.020512 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.004057 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MUCEN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.043948 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.028073 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
AMPH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.027659 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TQF7, Q8R1C4 | Gene names | Amph, Amph1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.028838 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PF21B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.031542 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.047560 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.026887 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
SEPT9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.019241 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.007854 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.057337 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
DSG2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.004284 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14126 | Gene names | DSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor (HDGC). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.030944 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.010365 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
NUP98_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.038919 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.010972 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
TACT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.037544 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P40200 | Gene names | CD96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell surface protein tactile precursor (T cell-activated increased late expression protein) (CD96 antigen). | |||||
|
TIP60_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.021353 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92993, O95624, Q13430, Q6GSE8, Q9BWK7 | Gene names | HTATIP, TIP60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat interactive protein) (cPLA(2)-interacting protein). | |||||
|
TIP60_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.021343 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.079034 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
DC1L2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.019578 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.007380 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.009575 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.028931 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.031074 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
EAN57_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.027661 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
GLYG2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.018423 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15488, O15485, O15486, O15487, O15489, O15490 | Gene names | GYG2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycogenin-2 (EC 2.4.1.186) (GN-2) (GN2). | |||||
|
GP108_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.022549 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91WD0, Q8BXE9, Q925B1 | Gene names | Gpr108, Lustr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR108 precursor (Lung seven transmembrane receptor 2). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.029349 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.010257 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.025781 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.047670 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PAPD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.052802 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0D3, Q3UXJ1, Q8C651 | Gene names | Papd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) RNA polymerase, mitochondrial precursor (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase) (PAP-associated domain-containing protein 1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.049746 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.024961 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
RFIP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.014413 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.016883 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.035129 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
A2BP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.010334 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NWB1, Q9NS20 | Gene names | A2BP1, A2BP | |||
|
Domain Architecture |
|
|||||
| Description | Ataxin-2-binding protein 1. | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.021597 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
FOXK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.006834 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.031474 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
LRC27_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.017188 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
MARK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.001626 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
ULK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.000648 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.015452 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CN145_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.008355 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
GABR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.030859 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.026341 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
K0310_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.029502 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.026989 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PP14C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.021748 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAE6, Q5VY83, Q96BB1, Q9H277 | Gene names | PPP1R14C, KEPI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14C (PKC-potentiated PP1 inhibitory protein) (Kinase-enhanced PP1 inhibitor) (Serologically defined breast cancer antigen NY-BR-81). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.029549 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
UBIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.009739 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.027622 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.032217 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
DPOLQ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.011759 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75417, O95160 | Gene names | POLQ, POLH | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase theta (EC 2.7.7.7) (DNA polymerase eta). | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.000313 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
GIDRP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.010968 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.006292 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.020729 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
IF16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.012158 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
K0090_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.045663 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N766, Q14700, Q63HL0, Q63HL3, Q8NBH8 | Gene names | KIAA0090 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0090 precursor. | |||||
|
K0090_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.045669 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C7X2, Q3TCG4, Q6ZQJ4, Q8K3W8 | Gene names | Kiaa0090 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0090 precursor. | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.029016 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.014432 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
SP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.001185 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
VRK3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.012284 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3G5, Q921W6 | Gene names | Vrk3 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase VRK3 (EC 2.7.11.1) (Vaccinia-related kinase 3). | |||||
|
ZN287_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | -0.000985 (rank : 121) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBT7 | Gene names | ZNF287 | |||
|
Domain Architecture |
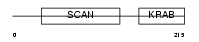
|
|||||
| Description | Zinc finger protein 287. | |||||
|
CD45_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.005014 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.004937 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
EPC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.020307 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2F5, Q5VW54, Q5VW56, Q5VW58, Q8NAQ4, Q8NE21, Q96LF4, Q96RR6, Q9H7T7 | Gene names | EPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 1. | |||||
|
GLI3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.000590 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
HBS1L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.009693 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q69ZS7, Q91Z32, Q9CVT2, Q9CZ95, Q9WTY5 | Gene names | Hbs1l, Hbs1, Kiaa1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein. | |||||
|
NOL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.021657 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60936, O60937 | Gene names | NOL3, ARC, NOP | |||
|
Domain Architecture |
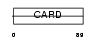
|
|||||
| Description | Nucleolar protein 3 (Apoptosis repressor with CARD) (Muscle-enriched cytoplasmic protein) (Myp) (Nucleolar protein of 30 kDa) (Nop30). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.011350 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
SP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.001403 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
UBAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.009436 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BH48, Q8BQ80, Q8BSW6, Q9D749, Q9ERV5 | Gene names | Ubap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 1 (UBAP) (Ubiquitin-associated protein NAG20). | |||||
|
UBIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.008795 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
|
ZCHC6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.036187 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
MUC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.054338 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.054975 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.073364 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.055110 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.062386 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.052543 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PAPOA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
PAPOA_HUMAN
|
||||||
| NC score | 0.987663 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
PAPOB_MOUSE
|
||||||
| NC score | 0.983091 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVP6, Q9R1R3 | Gene names | Papolb, Papt, Tpap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase beta (EC 2.7.7.19) (PAP beta) (Polynucleotide adenylyltransferase beta) (Testis-specific poly(A) polymerase). | |||||
|
PAPOB_HUMAN
|
||||||
| NC score | 0.981419 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRJ5, Q8NE14 | Gene names | PAPOLB, PAPT | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase beta (EC 2.7.7.19) (PAP beta) (Polynucleotide adenylyltransferase beta) (Testis-specific poly(A) polymerase). | |||||
|
PAPOG_HUMAN
|
||||||
| NC score | 0.972305 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BWT3, Q969N1, Q9H8L2, Q9HAD0 | Gene names | PAPOLG, PAP2, PAPG | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme) (Neo- poly(A) polymerase) (Neo-PAP). | |||||
|
PAPOG_MOUSE
|
||||||
| NC score | 0.967205 (rank : 6) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PCL9, Q8BZC9 | Gene names | Papolg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme). | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.117576 (rank : 7) | θ value | 0.000786445 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.088502 (rank : 8) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.085455 (rank : 9) | θ value | 0.0148317 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.082975 (rank : 10) | θ value | 0.0736092 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.079034 (rank : 11) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.073364 (rank : 12) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.069232 (rank : 13) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.067093 (rank : 14) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CD34_MOUSE
|
||||||
| NC score | 0.064268 (rank : 15) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64314, Q62550, Q62551 | Gene names | Cd34 | |||
|
Domain Architecture |
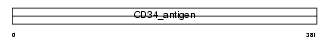
|
|||||
| Description | Hematopoietic progenitor cell antigen CD34 precursor. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.062386 (rank : 16) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.061470 (rank : 17) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.057676 (rank : 18) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.057337 (rank : 19) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.057149 (rank : 20) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
GA2L3_HUMAN
|
||||||
| NC score | 0.056873 (rank : 21) | θ value | 0.0431538 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.055110 (rank : 22) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.054975 (rank : 23) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.054338 (rank : 24) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.053973 (rank : 25) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PAPD1_MOUSE
|
||||||
| NC score | 0.052802 (rank : 26) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0D3, Q3UXJ1, Q8C651 | Gene names | Papd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) RNA polymerase, mitochondrial precursor (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase) (PAP-associated domain-containing protein 1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.052543 (rank : 27) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.049746 (rank : 28) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.047670 (rank : 29) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.047560 (rank : 30) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.045859 (rank : 31) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
K0090_MOUSE
|
||||||
| NC score | 0.045669 (rank : 32) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C7X2, Q3TCG4, Q6ZQJ4, Q8K3W8 | Gene names | Kiaa0090 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0090 precursor. | |||||
|
K0090_HUMAN
|
||||||
| NC score | 0.045663 (rank : 33) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N766, Q14700, Q63HL0, Q63HL3, Q8NBH8 | Gene names | KIAA0090 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0090 precursor. | |||||
|
MUCEN_MOUSE
|
||||||
| NC score | 0.043948 (rank : 34) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.039931 (rank : 35) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.039073 (rank : 36) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NUP98_HUMAN
|
||||||
| NC score | 0.038919 (rank : 37) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
TACT_HUMAN
|
||||||
| NC score | 0.037544 (rank : 38) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P40200 | Gene names | CD96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell surface protein tactile precursor (T cell-activated increased late expression protein) (CD96 antigen). | |||||
|
ZCHC6_MOUSE
|
||||||
| NC score | 0.036187 (rank : 39) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.036085 (rank : 40) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.035129 (rank : 41) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
SYNP2_HUMAN
|
||||||
| NC score | 0.034827 (rank : 42) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.033947 (rank : 43) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.033474 (rank : 44) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.032217 (rank : 45) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.032180 (rank : 46) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.031542 (rank : 47) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.031474 (rank : 48) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.031074 (rank : 49) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.030944 (rank : 50) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.030859 (rank : 51) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.029549 (rank : 52) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.029502 (rank : 53) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.029349 (rank : 54) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.029016 (rank : 55) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.028931 (rank : 56) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.028838 (rank : 57) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
TAF9B_HUMAN
|
||||||
| NC score | 0.028783 (rank : 58) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HBM6, Q9Y2S3 | Gene names | TAF9B, TAF9L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 9B (Transcription initiation factor TFIID subunit 9-like protein) (Transcription- associated factor TAFII31L) (Neuronal cell death-related protein 7) (DN-7). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.028073 (rank : 59) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.027969 (rank : 60) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
EAN57_HUMAN
|
||||||
| NC score | 0.027661 (rank : 61) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
AMPH_MOUSE
|
||||||
| NC score | 0.027659 (rank : 62) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TQF7, Q8R1C4 | Gene names | Amph, Amph1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.027622 (rank : 63) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.026989 (rank : 64) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.026887 (rank : 65) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
LRC27_MOUSE
|
||||||
| NC score | 0.026618 (rank : 66) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80YS5, Q9D6Z2 | Gene names | Lrrc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.026341 (rank : 67) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.025781 (rank : 68) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.024961 (rank : 69) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.023457 (rank : 70) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
GP108_MOUSE
|
||||||
| NC score | 0.022549 (rank : 71) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91WD0, Q8BXE9, Q925B1 | Gene names | Gpr108, Lustr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR108 precursor (Lung seven transmembrane receptor 2). | |||||
|
DAF1_MOUSE
|
||||||
| NC score | 0.021779 (rank : 72) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
PP14C_HUMAN
|
||||||
| NC score | 0.021748 (rank : 73) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAE6, Q5VY83, Q96BB1, Q9H277 | Gene names | PPP1R14C, KEPI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14C (PKC-potentiated PP1 inhibitory protein) (Kinase-enhanced PP1 inhibitor) (Serologically defined breast cancer antigen NY-BR-81). | |||||
|
NOL3_HUMAN
|
||||||
| NC score | 0.021657 (rank : 74) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60936, O60937 | Gene names | NOL3, ARC, NOP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein 3 (Apoptosis repressor with CARD) (Muscle-enriched cytoplasmic protein) (Myp) (Nucleolar protein of 30 kDa) (Nop30). | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.021597 (rank : 75) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
TIP60_HUMAN
|
||||||
| NC score | 0.021353 (rank : 76) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92993, O95624, Q13430, Q6GSE8, Q9BWK7 | Gene names | HTATIP, TIP60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat interactive protein) (cPLA(2)-interacting protein). | |||||
|
TIP60_MOUSE
|
||||||
| NC score | 0.021343 (rank : 77) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.020729 (rank : 78) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.020512 (rank : 79) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
EPC1_HUMAN
|
||||||
| NC score | 0.020307 (rank : 80) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2F5, Q5VW54, Q5VW56, Q5VW58, Q8NAQ4, Q8NE21, Q96LF4, Q96RR6, Q9H7T7 | Gene names | EPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 1. | |||||
|
DC1L2_MOUSE
|
||||||
| NC score | 0.019578 (rank : 81) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
SEPT9_MOUSE
|
||||||
| NC score | 0.019241 (rank : 82) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
GLYG2_HUMAN
|
||||||
| NC score | 0.018423 (rank : 83) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15488, O15485, O15486, O15487, O15489, O15490 | Gene names | GYG2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycogenin-2 (EC 2.4.1.186) (GN-2) (GN2). | |||||
|
LRC27_HUMAN
|
||||||
| NC score | 0.017188 (rank : 84) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.016883 (rank : 85) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.015452 (rank : 86) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.014432 (rank : 87) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RFIP1_MOUSE
|
||||||
| NC score | 0.014413 (rank : 88) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.013671 (rank : 89) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
VRK3_MOUSE
|
||||||
| NC score | 0.012284 (rank : 90) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3G5, Q921W6 | Gene names | Vrk3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase VRK3 (EC 2.7.11.1) (Vaccinia-related kinase 3). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.012158 (rank : 91) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
DPOLQ_HUMAN
|
||||||
| NC score | 0.011759 (rank : 92) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75417, O95160 | Gene names | POLQ, POLH | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase theta (EC 2.7.7.7) (DNA polymerase eta). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.011350 (rank : 93) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.010972 (rank : 94) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
GIDRP_MOUSE
|
||||||
| NC score | 0.010968 (rank : 95) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.010365 (rank : 96) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
A2BP1_HUMAN
|
||||||
| NC score | 0.010334 (rank : 97) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NWB1, Q9NS20 | Gene names | A2BP1, A2BP | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-2-binding protein 1. | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.010257 (rank : 98) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
UBIP1_MOUSE
|
||||||
| NC score | 0.009739 (rank : 99) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
HBS1L_MOUSE
|
||||||
| NC score | 0.009693 (rank : 100) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q69ZS7, Q91Z32, Q9CVT2, Q9CZ95, Q9WTY5 | Gene names | Hbs1l, Hbs1, Kiaa1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein. | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.009575 (rank : 101) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
UBAP1_MOUSE
|
||||||
| NC score | 0.009436 (rank : 102) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BH48, Q8BQ80, Q8BSW6, Q9D749, Q9ERV5 | Gene names | Ubap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 1 (UBAP) (Ubiquitin-associated protein NAG20). | |||||
|
UBIP1_HUMAN
|
||||||
| NC score | 0.008795 (rank : 103) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.008355 (rank : 104) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
RXRA_HUMAN
|
||||||
| NC score | 0.008343 (rank : 105) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.007854 (rank : 106) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.007380 (rank : 107) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
FOXK2_HUMAN
|
||||||
| NC score | 0.006834 (rank : 108) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.006292 (rank : 109) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.005014 (rank : 110) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.004937 (rank : 111) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
ZBT38_HUMAN
|
||||||
| NC score | 0.004532 (rank : 112) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
DSG2_HUMAN
|
||||||
| NC score | 0.004284 (rank : 113) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14126 | Gene names | DSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor (HDGC). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.004057 (rank : 114) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MARK2_MOUSE
|
||||||
| NC score | 0.001626 (rank : 115) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.001403 (rank : 116) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.001185 (rank : 117) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
ULK2_HUMAN
|
||||||
| NC score | 0.000648 (rank : 118) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
GLI3_HUMAN
|
||||||
| NC score | 0.000590 (rank : 119) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
EVI1_HUMAN
|
||||||
| NC score | 0.000313 (rank : 120) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
ZN287_HUMAN
|
||||||
| NC score | -0.000985 (rank : 121) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBT7 | Gene names | ZNF287 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 287. | |||||