Please be patient as the page loads
|
HBS1L_MOUSE
|
||||||
| SwissProt Accessions | Q69ZS7, Q91Z32, Q9CVT2, Q9CZ95, Q9WTY5 | Gene names | Hbs1l, Hbs1, Kiaa1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HBS1L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992304 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y450, Q5T7G3, Q8NDW9, Q9UPW3 | Gene names | HBS1L, HBS1, KIAA1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein (ERFS). | |||||
|
HBS1L_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q69ZS7, Q91Z32, Q9CVT2, Q9CZ95, Q9WTY5 | Gene names | Hbs1l, Hbs1, Kiaa1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein. | |||||
|
GSPT1_HUMAN
|
||||||
| θ value | 3.33315e-87 (rank : 3) | NC score | 0.953480 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15170 | Gene names | GSPT1 | |||
|
Domain Architecture |

|
|||||
| Description | G1 to S phase transition protein 1 homolog (GTP-binding protein GST1- HS). | |||||
|
EF1A2_HUMAN
|
||||||
| θ value | 5.6855e-87 (rank : 4) | NC score | 0.944563 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05639, P54266 | Gene names | EEF1A2, EEF1AL, STN | |||
|
Domain Architecture |
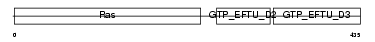
|
|||||
| Description | Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1). | |||||
|
EF1A2_MOUSE
|
||||||
| θ value | 5.6855e-87 (rank : 5) | NC score | 0.944564 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P62631, P27706 | Gene names | Eef1a2, Eef1al, Stn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1). | |||||
|
GSPT1_MOUSE
|
||||||
| θ value | 6.28603e-86 (rank : 6) | NC score | 0.952542 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R050, O88179 | Gene names | Gspt1 | |||
|
Domain Architecture |

|
|||||
| Description | G1 to S phase transition protein 1 homolog. | |||||
|
EF1A1_HUMAN
|
||||||
| θ value | 1.18549e-84 (rank : 7) | NC score | 0.944411 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P68104, P04719, P04720 | Gene names | EEF1A1, EEF1A, EF1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu). | |||||
|
EF1A1_MOUSE
|
||||||
| θ value | 1.18549e-84 (rank : 8) | NC score | 0.944522 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10126, Q61511, Q6ZWN2, Q8BMB8, Q8BVS8, Q99KU5 | Gene names | Eef1a1, Eef1a | |||
|
Domain Architecture |
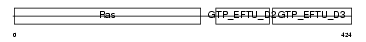
|
|||||
| Description | Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu). | |||||
|
EFTU_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 9) | NC score | 0.833751 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P49411, O15276 | Gene names | TUFM | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor Tu, mitochondrial precursor (EF-Tu) (P43). | |||||
|
EFTU_MOUSE
|
||||||
| θ value | 1.28732e-30 (rank : 10) | NC score | 0.834716 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BFR5, Q6P919 | Gene names | Tufm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor Tu, mitochondrial precursor. | |||||
|
GTPB1_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 11) | NC score | 0.600651 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00178, Q6IC67 | Gene names | GTPBP1 | |||
|
Domain Architecture |
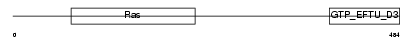
|
|||||
| Description | GTP-binding protein 1 (G-protein 1) (GP-1) (GP1). | |||||
|
GTPB1_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 12) | NC score | 0.596972 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08582, Q3UD96, Q3UGW6, Q545R1, Q80ZY5 | Gene names | Gtpbp1, Gtpbp | |||
|
Domain Architecture |
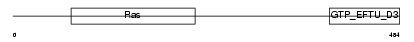
|
|||||
| Description | GTP-binding protein 1 (G-protein 1) (GP-1) (GP1). | |||||
|
GTPB2_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 13) | NC score | 0.551103 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BX10, Q5T7E8, Q8ND84, Q8TAH7, Q8WUA5, Q9HCS9, Q9NRU4, Q9NX60 | Gene names | GTPBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2. | |||||
|
GTPB2_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 14) | NC score | 0.551087 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3UJK4, Q9EST7, Q9JIX7 | Gene names | Gtpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2 (GTP-binding-like protein 2). | |||||
|
SELB_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 15) | NC score | 0.630904 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JHW4 | Gene names | Eefsec, Selb | |||
|
Domain Architecture |

|
|||||
| Description | Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific) (mSelB). | |||||
|
SELB_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 16) | NC score | 0.611667 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P57772 | Gene names | EEFSEC, SELB | |||
|
Domain Architecture |
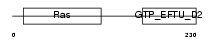
|
|||||
| Description | Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific). | |||||
|
IF2M_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 17) | NC score | 0.448382 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46199 | Gene names | MTIF2 | |||
|
Domain Architecture |
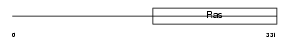
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
EFG1_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 18) | NC score | 0.381231 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96RP9, Q96T39 | Gene names | GFM1, EFG, EFG1, GFM | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 1, mitochondrial precursor (mEF-G 1) (Elongation factor G1). | |||||
|
EFG1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 19) | NC score | 0.369441 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K0D5, Q921D6, Q924I0 | Gene names | Gfm1, Efg, Efg1, Gfm | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 1, mitochondrial precursor (mEF-G 1) (Elongation factor G1). | |||||
|
EFG2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 20) | NC score | 0.379358 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q969S9, Q8WYI0 | Gene names | GFM2, EFG2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 2, mitochondrial precursor (mEF-G 2) (Elongation factor G2). | |||||
|
EFG2_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 21) | NC score | 0.365592 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R2Q4 | Gene names | Gfm2, Efg2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 2, mitochondrial precursor (mEF-G 2) (Elongation factor G2). | |||||
|
IF2M_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 22) | NC score | 0.367372 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |
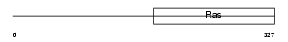
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
IF2G_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 23) | NC score | 0.354576 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41091 | Gene names | EIF2S3, EIF2G | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3 (Eukaryotic translation initiation factor 2 subunit gamma) (eIF-2-gamma). | |||||
|
IF2G_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 24) | NC score | 0.354567 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0N1 | Gene names | Eif2s3x | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3, X-linked (Eukaryotic translation initiation factor 2 subunit gamma, X-linked) (eIF-2-gamma X). | |||||
|
IF2H_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 25) | NC score | 0.356310 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z0N2 | Gene names | Eif2s3y | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3, Y-linked (Eukaryotic translation initiation factor 2 subunit gamma, Y-linked) (eIF-2-gamma Y). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 26) | NC score | 0.144776 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
BAIP2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.054333 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQB8, O43858, Q53HB1, Q86WC1, Q8N5C0, Q96CR7, Q9UBR3, Q9UQ43 | Gene names | BAIAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2 (BAI1- associated protein 2) (BAI-associated protein 2) (Protein BAP2) (Insulin receptor substrate p53) (IRSp53) (Insulin receptor substrate protein of 53 kDa) (Insulin receptor substrate p53/p58) (IRSp53/58) (IRS-58) (Fas ligand-associated factor 3) (FLAF3). | |||||
|
EF2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.239846 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13639 | Gene names | EEF2, EF2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 (EF-2). | |||||
|
EF2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.242122 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P58252, Q544E4 | Gene names | Eef2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 (EF-2). | |||||
|
U5S1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 30) | NC score | 0.195174 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15029, Q9BUR0 | Gene names | EFTUD2, KIAA0031, SNRP116 | |||
|
Domain Architecture |

|
|||||
| Description | 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP- specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP- binding domain protein 2) (hSNU114). | |||||
|
U5S1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 31) | NC score | 0.195103 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08810 | Gene names | Eftud2, Snrp116 | |||
|
Domain Architecture |

|
|||||
| Description | 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP- specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP- binding domain protein 2). | |||||
|
APC_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.026330 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
BAIP2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.050858 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BKX1, Q91V97, Q923V9, Q923W0 | Gene names | Baiap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2 (BAI1- associated protein 2) (BAI-associated protein 2) (Insulin receptor substrate p53) (IRSp53) (Insulin receptor substrate protein of 53 kDa) (Insulin receptor tyrosine kinase 53 kDa substrate). | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.006941 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.021396 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
SH2D3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.018121 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZS8, Q9JME1 | Gene names | Sh2d3c, Chat, Shep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (SH2 domain-containing Eph receptor- binding protein 1) (Cas/HEF1-associated signal transducer). | |||||
|
SERA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.027062 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43175, Q5SZU3, Q9BQ01 | Gene names | PHGDH, PGDH3 | |||
|
Domain Architecture |
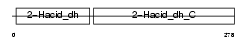
|
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH). | |||||
|
SMDF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.024427 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15491 | Gene names | NRG1, GGF, HGL, HRGA, NDF, SMDF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuregulin-1, sensory and motor neuron-derived factor isoform. | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.010575 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.004160 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
ODP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.015158 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.009157 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
PAPOA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.011550 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
SERA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.020652 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61753, Q75SV9 | Gene names | Phgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH) (A10). | |||||
|
ABCF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.015452 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UG63, O60864, Q96TE8 | Gene names | ABCF2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family F member 2 (Iron-inhibited ABC transporter 2). | |||||
|
ABCF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.015489 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99LE6, Q3UA24, Q8C1S5, Q9JL48 | Gene names | Abcf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 2. | |||||
|
JPH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.007064 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.007537 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.005988 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.000840 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.011365 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | -0.001988 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.012962 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CJ047_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.010910 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
DAAM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.004580 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | -0.002338 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.007456 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.013537 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.012717 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.012213 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
DIP2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.006489 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14689, Q8IVA3, Q8N4S2, Q8TD89, Q96ML9 | Gene names | DIP2A, C21orf106, DIP2, KIAA0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
HMCN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | -0.001927 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.006394 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PAPOA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.009693 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
TFE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.005665 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
HBS1L_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q69ZS7, Q91Z32, Q9CVT2, Q9CZ95, Q9WTY5 | Gene names | Hbs1l, Hbs1, Kiaa1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein. | |||||
|
HBS1L_HUMAN
|
||||||
| NC score | 0.992304 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y450, Q5T7G3, Q8NDW9, Q9UPW3 | Gene names | HBS1L, HBS1, KIAA1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein (ERFS). | |||||
|
GSPT1_HUMAN
|
||||||
| NC score | 0.953480 (rank : 3) | θ value | 3.33315e-87 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15170 | Gene names | GSPT1 | |||
|
Domain Architecture |

|
|||||
| Description | G1 to S phase transition protein 1 homolog (GTP-binding protein GST1- HS). | |||||
|
GSPT1_MOUSE
|
||||||
| NC score | 0.952542 (rank : 4) | θ value | 6.28603e-86 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R050, O88179 | Gene names | Gspt1 | |||
|
Domain Architecture |

|
|||||
| Description | G1 to S phase transition protein 1 homolog. | |||||
|
EF1A2_MOUSE
|
||||||
| NC score | 0.944564 (rank : 5) | θ value | 5.6855e-87 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P62631, P27706 | Gene names | Eef1a2, Eef1al, Stn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1). | |||||
|
EF1A2_HUMAN
|
||||||
| NC score | 0.944563 (rank : 6) | θ value | 5.6855e-87 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05639, P54266 | Gene names | EEF1A2, EEF1AL, STN | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1). | |||||
|
EF1A1_MOUSE
|
||||||
| NC score | 0.944522 (rank : 7) | θ value | 1.18549e-84 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10126, Q61511, Q6ZWN2, Q8BMB8, Q8BVS8, Q99KU5 | Gene names | Eef1a1, Eef1a | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu). | |||||
|
EF1A1_HUMAN
|
||||||
| NC score | 0.944411 (rank : 8) | θ value | 1.18549e-84 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P68104, P04719, P04720 | Gene names | EEF1A1, EEF1A, EF1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu). | |||||
|
EFTU_MOUSE
|
||||||
| NC score | 0.834716 (rank : 9) | θ value | 1.28732e-30 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BFR5, Q6P919 | Gene names | Tufm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor Tu, mitochondrial precursor. | |||||
|
EFTU_HUMAN
|
||||||
| NC score | 0.833751 (rank : 10) | θ value | 1.28732e-30 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P49411, O15276 | Gene names | TUFM | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor Tu, mitochondrial precursor (EF-Tu) (P43). | |||||
|
SELB_MOUSE
|
||||||
| NC score | 0.630904 (rank : 11) | θ value | 6.00763e-12 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JHW4 | Gene names | Eefsec, Selb | |||
|
Domain Architecture |

|
|||||
| Description | Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific) (mSelB). | |||||
|
SELB_HUMAN
|
||||||
| NC score | 0.611667 (rank : 12) | θ value | 3.29651e-10 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P57772 | Gene names | EEFSEC, SELB | |||
|
Domain Architecture |

|
|||||
| Description | Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific). | |||||
|
GTPB1_HUMAN
|
||||||
| NC score | 0.600651 (rank : 13) | θ value | 4.16044e-13 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00178, Q6IC67 | Gene names | GTPBP1 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein 1 (G-protein 1) (GP-1) (GP1). | |||||
|
GTPB1_MOUSE
|
||||||
| NC score | 0.596972 (rank : 14) | θ value | 1.2105e-12 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08582, Q3UD96, Q3UGW6, Q545R1, Q80ZY5 | Gene names | Gtpbp1, Gtpbp | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein 1 (G-protein 1) (GP-1) (GP1). | |||||
|
GTPB2_HUMAN
|
||||||
| NC score | 0.551103 (rank : 15) | θ value | 2.69671e-12 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BX10, Q5T7E8, Q8ND84, Q8TAH7, Q8WUA5, Q9HCS9, Q9NRU4, Q9NX60 | Gene names | GTPBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2. | |||||
|
GTPB2_MOUSE
|
||||||
| NC score | 0.551087 (rank : 16) | θ value | 2.69671e-12 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3UJK4, Q9EST7, Q9JIX7 | Gene names | Gtpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2 (GTP-binding-like protein 2). | |||||
|
IF2M_HUMAN
|
||||||
| NC score | 0.448382 (rank : 17) | θ value | 4.76016e-09 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46199 | Gene names | MTIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
EFG1_HUMAN
|
||||||
| NC score | 0.381231 (rank : 18) | θ value | 6.43352e-06 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96RP9, Q96T39 | Gene names | GFM1, EFG, EFG1, GFM | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 1, mitochondrial precursor (mEF-G 1) (Elongation factor G1). | |||||
|
EFG2_HUMAN
|
||||||
| NC score | 0.379358 (rank : 19) | θ value | 2.44474e-05 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q969S9, Q8WYI0 | Gene names | GFM2, EFG2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 2, mitochondrial precursor (mEF-G 2) (Elongation factor G2). | |||||
|
EFG1_MOUSE
|
||||||
| NC score | 0.369441 (rank : 20) | θ value | 1.87187e-05 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K0D5, Q921D6, Q924I0 | Gene names | Gfm1, Efg, Efg1, Gfm | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 1, mitochondrial precursor (mEF-G 1) (Elongation factor G1). | |||||
|
IF2M_MOUSE
|
||||||
| NC score | 0.367372 (rank : 21) | θ value | 0.00175202 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
EFG2_MOUSE
|
||||||
| NC score | 0.365592 (rank : 22) | θ value | 0.000121331 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R2Q4 | Gene names | Gfm2, Efg2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 2, mitochondrial precursor (mEF-G 2) (Elongation factor G2). | |||||
|
IF2H_MOUSE
|
||||||
| NC score | 0.356310 (rank : 23) | θ value | 0.00390308 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z0N2 | Gene names | Eif2s3y | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3, Y-linked (Eukaryotic translation initiation factor 2 subunit gamma, Y-linked) (eIF-2-gamma Y). | |||||
|
IF2G_HUMAN
|
||||||
| NC score | 0.354576 (rank : 24) | θ value | 0.00390308 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41091 | Gene names | EIF2S3, EIF2G | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3 (Eukaryotic translation initiation factor 2 subunit gamma) (eIF-2-gamma). | |||||
|
IF2G_MOUSE
|
||||||
| NC score | 0.354567 (rank : 25) | θ value | 0.00390308 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0N1 | Gene names | Eif2s3x | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3, X-linked (Eukaryotic translation initiation factor 2 subunit gamma, X-linked) (eIF-2-gamma X). | |||||
|
EF2_MOUSE
|
||||||
| NC score | 0.242122 (rank : 26) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P58252, Q544E4 | Gene names | Eef2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 (EF-2). | |||||
|
EF2_HUMAN
|
||||||
| NC score | 0.239846 (rank : 27) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13639 | Gene names | EEF2, EF2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 (EF-2). | |||||
|
U5S1_HUMAN
|
||||||
| NC score | 0.195174 (rank : 28) | θ value | 0.125558 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15029, Q9BUR0 | Gene names | EFTUD2, KIAA0031, SNRP116 | |||
|
Domain Architecture |

|
|||||
| Description | 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP- specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP- binding domain protein 2) (hSNU114). | |||||
|
U5S1_MOUSE
|
||||||
| NC score | 0.195103 (rank : 29) | θ value | 0.125558 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08810 | Gene names | Eftud2, Snrp116 | |||
|
Domain Architecture |

|
|||||
| Description | 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP- specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP- binding domain protein 2). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.144776 (rank : 30) | θ value | 0.0148317 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
BAIP2_HUMAN
|
||||||
| NC score | 0.054333 (rank : 31) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQB8, O43858, Q53HB1, Q86WC1, Q8N5C0, Q96CR7, Q9UBR3, Q9UQ43 | Gene names | BAIAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2 (BAI1- associated protein 2) (BAI-associated protein 2) (Protein BAP2) (Insulin receptor substrate p53) (IRSp53) (Insulin receptor substrate protein of 53 kDa) (Insulin receptor substrate p53/p58) (IRSp53/58) (IRS-58) (Fas ligand-associated factor 3) (FLAF3). | |||||
|
BAIP2_MOUSE
|
||||||
| NC score | 0.050858 (rank : 32) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BKX1, Q91V97, Q923V9, Q923W0 | Gene names | Baiap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2 (BAI1- associated protein 2) (BAI-associated protein 2) (Insulin receptor substrate p53) (IRSp53) (Insulin receptor substrate protein of 53 kDa) (Insulin receptor tyrosine kinase 53 kDa substrate). | |||||
|
SERA_HUMAN
|
||||||
| NC score | 0.027062 (rank : 33) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43175, Q5SZU3, Q9BQ01 | Gene names | PHGDH, PGDH3 | |||
|
Domain Architecture |

|
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.026330 (rank : 34) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
SMDF_HUMAN
|
||||||
| NC score | 0.024427 (rank : 35) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15491 | Gene names | NRG1, GGF, HGL, HRGA, NDF, SMDF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuregulin-1, sensory and motor neuron-derived factor isoform. | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.021396 (rank : 36) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
SERA_MOUSE
|
||||||
| NC score | 0.020652 (rank : 37) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61753, Q75SV9 | Gene names | Phgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH) (A10). | |||||
|
SH2D3_MOUSE
|
||||||
| NC score | 0.018121 (rank : 38) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZS8, Q9JME1 | Gene names | Sh2d3c, Chat, Shep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (SH2 domain-containing Eph receptor- binding protein 1) (Cas/HEF1-associated signal transducer). | |||||
|
ABCF2_MOUSE
|
||||||
| NC score | 0.015489 (rank : 39) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99LE6, Q3UA24, Q8C1S5, Q9JL48 | Gene names | Abcf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 2. | |||||
|
ABCF2_HUMAN
|
||||||
| NC score | 0.015452 (rank : 40) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UG63, O60864, Q96TE8 | Gene names | ABCF2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family F member 2 (Iron-inhibited ABC transporter 2). | |||||
|
ODP2_HUMAN
|
||||||
| NC score | 0.015158 (rank : 41) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.013537 (rank : 42) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.012962 (rank : 43) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.012717 (rank : 44) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.012213 (rank : 45) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
PAPOA_HUMAN
|
||||||
| NC score | 0.011550 (rank : 46) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.011365 (rank : 47) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
CJ047_HUMAN
|
||||||
| NC score | 0.010910 (rank : 48) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.010575 (rank : 49) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
PAPOA_MOUSE
|
||||||
| NC score | 0.009693 (rank : 50) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.009157 (rank : 51) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.007537 (rank : 52) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.007456 (rank : 53) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
JPH3_HUMAN
|
||||||
| NC score | 0.007064 (rank : 54) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.006941 (rank : 55) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
DIP2A_HUMAN
|
||||||
| NC score | 0.006489 (rank : 56) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14689, Q8IVA3, Q8N4S2, Q8TD89, Q96ML9 | Gene names | DIP2A, C21orf106, DIP2, KIAA0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.006394 (rank : 57) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.005988 (rank : 58) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TFE2_MOUSE
|
||||||
| NC score | 0.005665 (rank : 59) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
DAAM1_MOUSE
|
||||||
| NC score | 0.004580 (rank : 60) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.004160 (rank : 61) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.000840 (rank : 62) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
HMCN1_HUMAN
|
||||||
| NC score | -0.001927 (rank : 63) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | -0.001988 (rank : 64) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
EVI1_HUMAN
|
||||||
| NC score | -0.002338 (rank : 65) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||