Please be patient as the page loads
|
GSPT1_MOUSE
|
||||||
| SwissProt Accessions | Q8R050, O88179 | Gene names | Gspt1 | |||
|
Domain Architecture |

|
|||||
| Description | G1 to S phase transition protein 1 homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GSPT1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999398 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15170 | Gene names | GSPT1 | |||
|
Domain Architecture |

|
|||||
| Description | G1 to S phase transition protein 1 homolog (GTP-binding protein GST1- HS). | |||||
|
GSPT1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R050, O88179 | Gene names | Gspt1 | |||
|
Domain Architecture |

|
|||||
| Description | G1 to S phase transition protein 1 homolog. | |||||
|
HBS1L_HUMAN
|
||||||
| θ value | 3.93732e-88 (rank : 3) | NC score | 0.950023 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y450, Q5T7G3, Q8NDW9, Q9UPW3 | Gene names | HBS1L, HBS1, KIAA1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein (ERFS). | |||||
|
HBS1L_MOUSE
|
||||||
| θ value | 6.28603e-86 (rank : 4) | NC score | 0.952542 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q69ZS7, Q91Z32, Q9CVT2, Q9CZ95, Q9WTY5 | Gene names | Hbs1l, Hbs1, Kiaa1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein. | |||||
|
EF1A2_HUMAN
|
||||||
| θ value | 2.23575e-83 (rank : 5) | NC score | 0.947670 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q05639, P54266 | Gene names | EEF1A2, EEF1AL, STN | |||
|
Domain Architecture |
|
|||||
| Description | Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1). | |||||
|
EF1A2_MOUSE
|
||||||
| θ value | 2.91998e-83 (rank : 6) | NC score | 0.947613 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62631, P27706 | Gene names | Eef1a2, Eef1al, Stn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1). | |||||
|
EF1A1_HUMAN
|
||||||
| θ value | 2.47191e-82 (rank : 7) | NC score | 0.948109 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P68104, P04719, P04720 | Gene names | EEF1A1, EEF1A, EF1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu). | |||||
|
EF1A1_MOUSE
|
||||||
| θ value | 2.47191e-82 (rank : 8) | NC score | 0.947971 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10126, Q61511, Q6ZWN2, Q8BMB8, Q8BVS8, Q99KU5 | Gene names | Eef1a1, Eef1a | |||
|
Domain Architecture |
|
|||||
| Description | Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu). | |||||
|
EFTU_MOUSE
|
||||||
| θ value | 1.62847e-25 (rank : 9) | NC score | 0.830703 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BFR5, Q6P919 | Gene names | Tufm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor Tu, mitochondrial precursor. | |||||
|
EFTU_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 10) | NC score | 0.828572 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49411, O15276 | Gene names | TUFM | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor Tu, mitochondrial precursor (EF-Tu) (P43). | |||||
|
GTPB1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 11) | NC score | 0.572298 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00178, Q6IC67 | Gene names | GTPBP1 | |||
|
Domain Architecture |
|
|||||
| Description | GTP-binding protein 1 (G-protein 1) (GP-1) (GP1). | |||||
|
GTPB1_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 12) | NC score | 0.568592 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08582, Q3UD96, Q3UGW6, Q545R1, Q80ZY5 | Gene names | Gtpbp1, Gtpbp | |||
|
Domain Architecture |
|
|||||
| Description | GTP-binding protein 1 (G-protein 1) (GP-1) (GP1). | |||||
|
GTPB2_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 13) | NC score | 0.508432 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BX10, Q5T7E8, Q8ND84, Q8TAH7, Q8WUA5, Q9HCS9, Q9NRU4, Q9NX60 | Gene names | GTPBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2. | |||||
|
GTPB2_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 14) | NC score | 0.508489 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UJK4, Q9EST7, Q9JIX7 | Gene names | Gtpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2 (GTP-binding-like protein 2). | |||||
|
IF2M_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 15) | NC score | 0.433397 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46199 | Gene names | MTIF2 | |||
|
Domain Architecture |
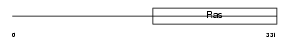
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
EFG1_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 16) | NC score | 0.374379 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96RP9, Q96T39 | Gene names | GFM1, EFG, EFG1, GFM | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 1, mitochondrial precursor (mEF-G 1) (Elongation factor G1). | |||||
|
IF2G_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 17) | NC score | 0.401663 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41091 | Gene names | EIF2S3, EIF2G | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3 (Eukaryotic translation initiation factor 2 subunit gamma) (eIF-2-gamma). | |||||
|
IF2G_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 18) | NC score | 0.401655 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0N1 | Gene names | Eif2s3x | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3, X-linked (Eukaryotic translation initiation factor 2 subunit gamma, X-linked) (eIF-2-gamma X). | |||||
|
IF2H_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 19) | NC score | 0.401348 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0N2 | Gene names | Eif2s3y | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3, Y-linked (Eukaryotic translation initiation factor 2 subunit gamma, Y-linked) (eIF-2-gamma Y). | |||||
|
SELB_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 20) | NC score | 0.585811 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P57772 | Gene names | EEFSEC, SELB | |||
|
Domain Architecture |
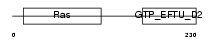
|
|||||
| Description | Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific). | |||||
|
SELB_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 21) | NC score | 0.600660 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHW4 | Gene names | Eefsec, Selb | |||
|
Domain Architecture |

|
|||||
| Description | Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific) (mSelB). | |||||
|
EFG1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 22) | NC score | 0.360795 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K0D5, Q921D6, Q924I0 | Gene names | Gfm1, Efg, Efg1, Gfm | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 1, mitochondrial precursor (mEF-G 1) (Elongation factor G1). | |||||
|
IF2M_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 23) | NC score | 0.361674 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |
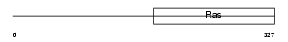
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
EFG2_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 24) | NC score | 0.348280 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R2Q4 | Gene names | Gfm2, Efg2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 2, mitochondrial precursor (mEF-G 2) (Elongation factor G2). | |||||
|
EFG2_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 25) | NC score | 0.359581 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q969S9, Q8WYI0 | Gene names | GFM2, EFG2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 2, mitochondrial precursor (mEF-G 2) (Elongation factor G2). | |||||
|
U5S1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.161339 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15029, Q9BUR0 | Gene names | EFTUD2, KIAA0031, SNRP116 | |||
|
Domain Architecture |

|
|||||
| Description | 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP- specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP- binding domain protein 2) (hSNU114). | |||||
|
U5S1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.161272 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O08810 | Gene names | Eftud2, Snrp116 | |||
|
Domain Architecture |

|
|||||
| Description | 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP- specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP- binding domain protein 2). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.124088 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
ROR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.002825 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01974, Q59GF5, Q5SPI5, Q9HAY7, Q9HB61 | Gene names | ROR2, NTRKR2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR2 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 2). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.015338 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.011971 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.023730 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.002401 (rank : 38) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
APBA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.004473 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.002817 (rank : 37) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.003454 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
EF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.199018 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13639 | Gene names | EEF2, EF2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 (EF-2). | |||||
|
EF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.201531 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P58252, Q544E4 | Gene names | Eef2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 (EF-2). | |||||
|
GSPT1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R050, O88179 | Gene names | Gspt1 | |||
|
Domain Architecture |

|
|||||
| Description | G1 to S phase transition protein 1 homolog. | |||||
|
GSPT1_HUMAN
|
||||||
| NC score | 0.999398 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15170 | Gene names | GSPT1 | |||
|
Domain Architecture |

|
|||||
| Description | G1 to S phase transition protein 1 homolog (GTP-binding protein GST1- HS). | |||||
|
HBS1L_MOUSE
|
||||||
| NC score | 0.952542 (rank : 3) | θ value | 6.28603e-86 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q69ZS7, Q91Z32, Q9CVT2, Q9CZ95, Q9WTY5 | Gene names | Hbs1l, Hbs1, Kiaa1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein. | |||||
|
HBS1L_HUMAN
|
||||||
| NC score | 0.950023 (rank : 4) | θ value | 3.93732e-88 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y450, Q5T7G3, Q8NDW9, Q9UPW3 | Gene names | HBS1L, HBS1, KIAA1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein (ERFS). | |||||
|
EF1A1_HUMAN
|
||||||
| NC score | 0.948109 (rank : 5) | θ value | 2.47191e-82 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P68104, P04719, P04720 | Gene names | EEF1A1, EEF1A, EF1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu). | |||||
|
EF1A1_MOUSE
|
||||||
| NC score | 0.947971 (rank : 6) | θ value | 2.47191e-82 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10126, Q61511, Q6ZWN2, Q8BMB8, Q8BVS8, Q99KU5 | Gene names | Eef1a1, Eef1a | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu). | |||||
|
EF1A2_HUMAN
|
||||||
| NC score | 0.947670 (rank : 7) | θ value | 2.23575e-83 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q05639, P54266 | Gene names | EEF1A2, EEF1AL, STN | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1). | |||||
|
EF1A2_MOUSE
|
||||||
| NC score | 0.947613 (rank : 8) | θ value | 2.91998e-83 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62631, P27706 | Gene names | Eef1a2, Eef1al, Stn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor 1-alpha 2 (EF-1-alpha-2) (Elongation factor 1 A-2) (eEF1A-2) (Statin S1). | |||||
|
EFTU_MOUSE
|
||||||
| NC score | 0.830703 (rank : 9) | θ value | 1.62847e-25 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BFR5, Q6P919 | Gene names | Tufm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elongation factor Tu, mitochondrial precursor. | |||||
|
EFTU_HUMAN
|
||||||
| NC score | 0.828572 (rank : 10) | θ value | 4.01107e-24 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49411, O15276 | Gene names | TUFM | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor Tu, mitochondrial precursor (EF-Tu) (P43). | |||||
|
SELB_MOUSE
|
||||||
| NC score | 0.600660 (rank : 11) | θ value | 6.43352e-06 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHW4 | Gene names | Eefsec, Selb | |||
|
Domain Architecture |

|
|||||
| Description | Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific) (mSelB). | |||||
|
SELB_HUMAN
|
||||||
| NC score | 0.585811 (rank : 12) | θ value | 4.92598e-06 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P57772 | Gene names | EEFSEC, SELB | |||
|
Domain Architecture |

|
|||||
| Description | Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific). | |||||
|
GTPB1_HUMAN
|
||||||
| NC score | 0.572298 (rank : 13) | θ value | 6.64225e-11 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00178, Q6IC67 | Gene names | GTPBP1 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein 1 (G-protein 1) (GP-1) (GP1). | |||||
|
GTPB1_MOUSE
|
||||||
| NC score | 0.568592 (rank : 14) | θ value | 1.133e-10 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08582, Q3UD96, Q3UGW6, Q545R1, Q80ZY5 | Gene names | Gtpbp1, Gtpbp | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein 1 (G-protein 1) (GP-1) (GP1). | |||||
|
GTPB2_MOUSE
|
||||||
| NC score | 0.508489 (rank : 15) | θ value | 4.45536e-07 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UJK4, Q9EST7, Q9JIX7 | Gene names | Gtpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2 (GTP-binding-like protein 2). | |||||
|
GTPB2_HUMAN
|
||||||
| NC score | 0.508432 (rank : 16) | θ value | 4.45536e-07 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BX10, Q5T7E8, Q8ND84, Q8TAH7, Q8WUA5, Q9HCS9, Q9NRU4, Q9NX60 | Gene names | GTPBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2. | |||||
|
IF2M_HUMAN
|
||||||
| NC score | 0.433397 (rank : 17) | θ value | 4.45536e-07 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46199 | Gene names | MTIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
IF2G_HUMAN
|
||||||
| NC score | 0.401663 (rank : 18) | θ value | 2.88788e-06 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41091 | Gene names | EIF2S3, EIF2G | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3 (Eukaryotic translation initiation factor 2 subunit gamma) (eIF-2-gamma). | |||||
|
IF2G_MOUSE
|
||||||
| NC score | 0.401655 (rank : 19) | θ value | 2.88788e-06 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0N1 | Gene names | Eif2s3x | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3, X-linked (Eukaryotic translation initiation factor 2 subunit gamma, X-linked) (eIF-2-gamma X). | |||||
|
IF2H_MOUSE
|
||||||
| NC score | 0.401348 (rank : 20) | θ value | 2.88788e-06 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0N2 | Gene names | Eif2s3y | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 3, Y-linked (Eukaryotic translation initiation factor 2 subunit gamma, Y-linked) (eIF-2-gamma Y). | |||||
|
EFG1_HUMAN
|
||||||
| NC score | 0.374379 (rank : 21) | θ value | 2.88788e-06 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96RP9, Q96T39 | Gene names | GFM1, EFG, EFG1, GFM | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 1, mitochondrial precursor (mEF-G 1) (Elongation factor G1). | |||||
|
IF2M_MOUSE
|
||||||
| NC score | 0.361674 (rank : 22) | θ value | 0.000158464 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
EFG1_MOUSE
|
||||||
| NC score | 0.360795 (rank : 23) | θ value | 1.43324e-05 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K0D5, Q921D6, Q924I0 | Gene names | Gfm1, Efg, Efg1, Gfm | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 1, mitochondrial precursor (mEF-G 1) (Elongation factor G1). | |||||
|
EFG2_HUMAN
|
||||||
| NC score | 0.359581 (rank : 24) | θ value | 0.00175202 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q969S9, Q8WYI0 | Gene names | GFM2, EFG2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 2, mitochondrial precursor (mEF-G 2) (Elongation factor G2). | |||||
|
EFG2_MOUSE
|
||||||
| NC score | 0.348280 (rank : 25) | θ value | 0.000786445 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R2Q4 | Gene names | Gfm2, Efg2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor G 2, mitochondrial precursor (mEF-G 2) (Elongation factor G2). | |||||
|
EF2_MOUSE
|
||||||
| NC score | 0.201531 (rank : 26) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P58252, Q544E4 | Gene names | Eef2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 (EF-2). | |||||
|
EF2_HUMAN
|
||||||
| NC score | 0.199018 (rank : 27) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13639 | Gene names | EEF2, EF2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 (EF-2). | |||||
|
U5S1_HUMAN
|
||||||
| NC score | 0.161339 (rank : 28) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15029, Q9BUR0 | Gene names | EFTUD2, KIAA0031, SNRP116 | |||
|
Domain Architecture |

|
|||||
| Description | 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP- specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP- binding domain protein 2) (hSNU114). | |||||
|
U5S1_MOUSE
|
||||||
| NC score | 0.161272 (rank : 29) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O08810 | Gene names | Eftud2, Snrp116 | |||
|
Domain Architecture |

|
|||||
| Description | 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP- specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP- binding domain protein 2). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.124088 (rank : 30) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.023730 (rank : 31) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.015338 (rank : 32) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.011971 (rank : 33) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
APBA3_HUMAN
|
||||||
| NC score | 0.004473 (rank : 34) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.003454 (rank : 35) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
ROR2_HUMAN
|
||||||
| NC score | 0.002825 (rank : 36) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01974, Q59GF5, Q5SPI5, Q9HAY7, Q9HB61 | Gene names | ROR2, NTRKR2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR2 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 2). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.002817 (rank : 37) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.002401 (rank : 38) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||