Please be patient as the page loads
|
MTFR1_MOUSE
|
||||||
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MTFR1_MOUSE
|
||||||
| θ value | 3.47146e-177 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
|
MTFR1_HUMAN
|
||||||
| θ value | 6.86991e-125 (rank : 2) | NC score | 0.982890 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15390, Q6IB94, Q86XH5, Q8IVD7 | Gene names | MTFR1, CHPPR, KIAA0009 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial fission regulator 1 (Chondrocyte protein with a poly- proline region). | |||||
|
FA54A_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 3) | NC score | 0.761785 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P444, Q5JWR7, Q6ZUE8, Q7L3U6, Q9BZ39 | Gene names | FAM54A, DUFD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
|
FA54A_MOUSE
|
||||||
| θ value | 6.18819e-25 (rank : 4) | NC score | 0.747277 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VED8 | Gene names | Fam54a, Dufd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 5) | NC score | 0.027861 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 0.47712 (rank : 6) | NC score | 0.016055 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
CORO6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.017583 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q920M5, Q920M3, Q920M4 | Gene names | Coro6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coronin-6 (Clipin E). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.024316 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.010717 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.018870 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
DCC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.022212 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.035956 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.025241 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.010255 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
FAK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.003231 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
NUP98_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.036257 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
ADCY3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.010947 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.019777 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
CADH4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.007484 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55283, Q2M208, Q5VZ44, Q9BZ05 | Gene names | CDH4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
ATF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.023390 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.023459 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
CNTN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.010481 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P232, Q9H039 | Gene names | CNTN3, KIAA1496, PANG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.007533 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
IKAR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.001550 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13422, O00598, Q8WVA3 | Gene names | IKZF1, IK1, IKAROS, LYF1, ZNFN1A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding protein Ikaros (IKAROS family zinc finger protein 1) (Lymphoid transcription factor LyF-1). | |||||
|
K1914_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.022130 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.011096 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
CADH4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.006757 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P39038 | Gene names | Cdh4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
LHX61_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.008389 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1R0, Q9R1R1 | Gene names | Lhx6.1 | |||
|
Domain Architecture |
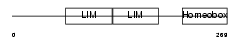
|
|||||
| Description | LIM/homeobox protein Lhx6.1. | |||||
|
LIRB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.008715 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.001888 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.021440 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.021814 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.007286 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.018600 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.007556 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.016831 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
LHX61_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.007571 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPM6, Q5T7S8, Q9NTK3, Q9UPM5 | Gene names | LHX6, LHX6.1 | |||
|
Domain Architecture |
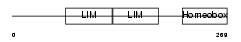
|
|||||
| Description | LIM/homeobox protein Lhx6.1 (Lhx6). | |||||
|
PAR3L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.009699 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
MTFR1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.47146e-177 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
|
MTFR1_HUMAN
|
||||||
| NC score | 0.982890 (rank : 2) | θ value | 6.86991e-125 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15390, Q6IB94, Q86XH5, Q8IVD7 | Gene names | MTFR1, CHPPR, KIAA0009 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial fission regulator 1 (Chondrocyte protein with a poly- proline region). | |||||
|
FA54A_HUMAN
|
||||||
| NC score | 0.761785 (rank : 3) | θ value | 2.12685e-25 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P444, Q5JWR7, Q6ZUE8, Q7L3U6, Q9BZ39 | Gene names | FAM54A, DUFD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
|
FA54A_MOUSE
|
||||||
| NC score | 0.747277 (rank : 4) | θ value | 6.18819e-25 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VED8 | Gene names | Fam54a, Dufd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
|
NUP98_HUMAN
|
||||||
| NC score | 0.036257 (rank : 5) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.035956 (rank : 6) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.027861 (rank : 7) | θ value | 0.47712 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.025241 (rank : 8) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.024316 (rank : 9) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.023459 (rank : 10) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.023390 (rank : 11) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
DCC_HUMAN
|
||||||
| NC score | 0.022212 (rank : 12) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
K1914_HUMAN
|
||||||
| NC score | 0.022130 (rank : 13) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.021814 (rank : 14) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.021440 (rank : 15) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.019777 (rank : 16) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.018870 (rank : 17) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.018600 (rank : 18) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
CORO6_MOUSE
|
||||||
| NC score | 0.017583 (rank : 19) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q920M5, Q920M3, Q920M4 | Gene names | Coro6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coronin-6 (Clipin E). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.016831 (rank : 20) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.016055 (rank : 21) | θ value | 0.47712 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.011096 (rank : 22) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ADCY3_HUMAN
|
||||||
| NC score | 0.010947 (rank : 23) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.010717 (rank : 24) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CNTN3_HUMAN
|
||||||
| NC score | 0.010481 (rank : 25) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P232, Q9H039 | Gene names | CNTN3, KIAA1496, PANG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.010255 (rank : 26) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
PAR3L_HUMAN
|
||||||
| NC score | 0.009699 (rank : 27) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
LIRB3_HUMAN
|
||||||
| NC score | 0.008715 (rank : 28) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
LHX61_MOUSE
|
||||||
| NC score | 0.008389 (rank : 29) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1R0, Q9R1R1 | Gene names | Lhx6.1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx6.1. | |||||
|
LHX61_HUMAN
|
||||||
| NC score | 0.007571 (rank : 30) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPM6, Q5T7S8, Q9NTK3, Q9UPM5 | Gene names | LHX6, LHX6.1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx6.1 (Lhx6). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.007556 (rank : 31) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.007533 (rank : 32) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CADH4_HUMAN
|
||||||
| NC score | 0.007484 (rank : 33) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55283, Q2M208, Q5VZ44, Q9BZ05 | Gene names | CDH4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.007286 (rank : 34) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CADH4_MOUSE
|
||||||
| NC score | 0.006757 (rank : 35) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P39038 | Gene names | Cdh4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
FAK2_MOUSE
|
||||||
| NC score | 0.003231 (rank : 36) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.001888 (rank : 37) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
IKAR_HUMAN
|
||||||
| NC score | 0.001550 (rank : 38) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13422, O00598, Q8WVA3 | Gene names | IKZF1, IK1, IKAROS, LYF1, ZNFN1A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding protein Ikaros (IKAROS family zinc finger protein 1) (Lymphoid transcription factor LyF-1). | |||||