Please be patient as the page loads
|
PACS1_MOUSE
|
||||||
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PACS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.988326 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6VY07, Q6PJY6, Q6PKB6, Q7Z590, Q7Z5W4, Q8N8K6, Q96MW0, Q9NW92, Q9ULP5 | Gene names | PACS1, KIAA1175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
PACS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 3) | NC score | 0.115728 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 4) | NC score | 0.059725 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 5) | NC score | 0.073910 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.072722 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 7) | NC score | 0.111413 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.100714 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.066026 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.067894 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.069170 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.023652 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.075833 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
TBX21_MOUSE
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.035794 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKD8, Q9R0A6 | Gene names | Tbx21, Tbet, Tblym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.080327 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.059709 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
JHD1B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.041071 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.040353 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.076378 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.047513 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.042918 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.022111 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.080348 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.035514 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.054270 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.012658 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.047245 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
K0240_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.033746 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.057834 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.031715 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.033624 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.054575 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
K1219_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.040784 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86X10, Q5TG31, Q8N3D1, Q8WWC0, Q9H3X8, Q9UJR1, Q9ULK1, Q9Y3G9 | Gene names | KIAA1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
PALM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.057660 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.035704 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
CJ055_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.054938 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SWW7, Q8NAK4 | Gene names | C10orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf55. | |||||
|
IPR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.044620 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.031378 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.022175 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
SI1L1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.023795 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.056684 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.003358 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ACD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.047566 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96AP0, Q562H5, Q9H8F9 | Gene names | ACD, PIP1, PTOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adrenocortical dysplasia protein homolog (POT1 and TIN2-interacting protein). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.053379 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.017652 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.024049 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.026199 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.046711 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.015949 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.024909 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
SLAF6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.030324 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96DU3, Q5TAT3, Q96DV0 | Gene names | SLAMF6, KALI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLAM family member 6 precursor (NK-T-B-antigen) (NTB-A) (Activating NK receptor). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.030404 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
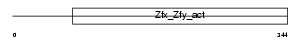
ZFA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.000391 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23607 | Gene names | Zfa | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger autosomal protein. | |||||
|
ESPL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.018629 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14674 | Gene names | ESPL1, ESP1, KIAA0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.011837 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
MAGAB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.014185 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |
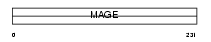
|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAML1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.022835 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.042650 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.011823 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
APC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.044734 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.026556 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.022403 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.015470 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.011589 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.008277 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.013355 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.053232 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.029303 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.033283 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.021110 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.011334 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
AHI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.008879 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
APC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.034758 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.027647 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CX033_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.016972 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UXV4, Q5H9D1 | Gene names | CXorf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf33 precursor. | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.010542 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
IF4B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.033102 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.019390 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.014172 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.030954 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.023229 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RAE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.010235 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24386, O43732 | Gene names | CHM, REP1, TCD | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein) (TCD protein). | |||||
|
TAF6L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.020091 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |
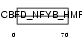
|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
TIF1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.007775 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TRA2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.008571 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.008571 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.014298 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050905 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
PACS1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
PACS1_HUMAN
|
||||||
| NC score | 0.988326 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6VY07, Q6PJY6, Q6PKB6, Q7Z590, Q7Z5W4, Q8N8K6, Q96MW0, Q9NW92, Q9ULP5 | Gene names | PACS1, KIAA1175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.115728 (rank : 3) | θ value | 0.00228821 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.111413 (rank : 4) | θ value | 0.0148317 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.100714 (rank : 5) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.080348 (rank : 6) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.080327 (rank : 7) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.076378 (rank : 8) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.075833 (rank : 9) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.073910 (rank : 10) | θ value | 0.00869519 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.072722 (rank : 11) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.069170 (rank : 12) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.067894 (rank : 13) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.066026 (rank : 14) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.059725 (rank : 15) | θ value | 0.00509761 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.059709 (rank : 16) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.057834 (rank : 17) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.057660 (rank : 18) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.056684 (rank : 19) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CJ055_HUMAN
|
||||||
| NC score | 0.054938 (rank : 20) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SWW7, Q8NAK4 | Gene names | C10orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf55. | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.054575 (rank : 21) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.054270 (rank : 22) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.053379 (rank : 23) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.053232 (rank : 24) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.050905 (rank : 25) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
ACD_HUMAN
|
||||||
| NC score | 0.047566 (rank : 26) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96AP0, Q562H5, Q9H8F9 | Gene names | ACD, PIP1, PTOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adrenocortical dysplasia protein homolog (POT1 and TIN2-interacting protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.047513 (rank : 27) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.047245 (rank : 28) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.046711 (rank : 29) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.044734 (rank : 30) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
IPR1_MOUSE
|
||||||
| NC score | 0.044620 (rank : 31) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.042918 (rank : 32) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.042650 (rank : 33) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
JHD1B_MOUSE
|
||||||
| NC score | 0.041071 (rank : 34) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
K1219_HUMAN
|
||||||
| NC score | 0.040784 (rank : 35) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86X10, Q5TG31, Q8N3D1, Q8WWC0, Q9H3X8, Q9UJR1, Q9ULK1, Q9Y3G9 | Gene names | KIAA1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.040353 (rank : 36) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
TBX21_MOUSE
|
||||||
| NC score | 0.035794 (rank : 37) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKD8, Q9R0A6 | Gene names | Tbx21, Tbet, Tblym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.035704 (rank : 38) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.035514 (rank : 39) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.034758 (rank : 40) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
K0240_HUMAN
|
||||||
| NC score | 0.033746 (rank : 41) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.033624 (rank : 42) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.033283 (rank : 43) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
IF4B_HUMAN
|
||||||
| NC score | 0.033102 (rank : 44) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.031715 (rank : 45) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.031378 (rank : 46) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.030954 (rank : 47) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.030404 (rank : 48) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
SLAF6_HUMAN
|
||||||
| NC score | 0.030324 (rank : 49) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96DU3, Q5TAT3, Q96DV0 | Gene names | SLAMF6, KALI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLAM family member 6 precursor (NK-T-B-antigen) (NTB-A) (Activating NK receptor). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.029303 (rank : 50) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.027647 (rank : 51) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.026556 (rank : 52) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.026199 (rank : 53) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.024909 (rank : 54) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.024049 (rank : 55) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SI1L1_MOUSE
|
||||||
| NC score | 0.023795 (rank : 56) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.023652 (rank : 57) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.023229 (rank : 58) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
MAML1_HUMAN
|
||||||
| NC score | 0.022835 (rank : 59) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.022403 (rank : 60) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.022175 (rank : 61) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.022111 (rank : 62) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.021110 (rank : 63) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
TAF6L_MOUSE
|
||||||
| NC score | 0.020091 (rank : 64) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |

|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.019390 (rank : 65) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ESPL1_HUMAN
|
||||||
| NC score | 0.018629 (rank : 66) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14674 | Gene names | ESPL1, ESP1, KIAA0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.017652 (rank : 67) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
CX033_HUMAN
|
||||||
| NC score | 0.016972 (rank : 68) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UXV4, Q5H9D1 | Gene names | CXorf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf33 precursor. | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.015949 (rank : 69) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.015470 (rank : 70) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.014298 (rank : 71) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
MAGAB_HUMAN
|
||||||
| NC score | 0.014185 (rank : 72) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.014172 (rank : 73) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.013355 (rank : 74) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.012658 (rank : 75) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.011837 (rank : 76) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.011823 (rank : 77) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.011589 (rank : 78) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.011334 (rank : 79) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.010542 (rank : 80) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
RAE1_HUMAN
|
||||||
| NC score | 0.010235 (rank : 81) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24386, O43732 | Gene names | CHM, REP1, TCD | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein) (TCD protein). | |||||
|
AHI1_HUMAN
|
||||||
| NC score | 0.008879 (rank : 82) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
TRA2B_HUMAN
|
||||||
| NC score | 0.008571 (rank : 83) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| NC score | 0.008571 (rank : 84) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.008277 (rank : 85) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
TIF1B_HUMAN
|
||||||
| NC score | 0.007775 (rank : 86) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.003358 (rank : 87) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ZFA_MOUSE
|
||||||
| NC score | 0.000391 (rank : 88) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23607 | Gene names | Zfa | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger autosomal protein. | |||||