Please be patient as the page loads
|
PKDRE_MOUSE
|
||||||
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PKDRE_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.984787 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 3) | NC score | 0.586976 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 4) | NC score | 0.574301 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 2.43343e-21 (rank : 5) | NC score | 0.579567 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 9.24701e-21 (rank : 6) | NC score | 0.577463 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 7) | NC score | 0.571020 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 8) | NC score | 0.324611 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 9) | NC score | 0.559714 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
LOX5_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 10) | NC score | 0.250935 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P09917 | Gene names | ALOX5, LOG5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
LOX5_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 11) | NC score | 0.240116 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48999 | Gene names | Alox5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
MCLN1_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 12) | NC score | 0.303518 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9GZU1, Q7Z4F7, Q9H292, Q9H4B3, Q9H4B5 | Gene names | MCOLN1, ML4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin) (MG-2). | |||||
|
MCLN1_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 13) | NC score | 0.296773 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99J21 | Gene names | Mcoln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin). | |||||
|
LX12B_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 14) | NC score | 0.177979 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75342 | Gene names | ALOX12B | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12R type (EC 1.13.11.-) (Epidermis-type lipoxygenase 12) (12R-lipoxygenase) (12R-LOX). | |||||
|
MCLN3_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 15) | NC score | 0.296263 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDD5, Q5T4H5, Q5T4H6, Q9NV09 | Gene names | MCOLN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN3_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 16) | NC score | 0.291543 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
LX12B_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 17) | NC score | 0.170147 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70582 | Gene names | Alox12b, Aloxe2 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12R type (EC 1.13.11.-) (Epidermis-type lipoxygenase 12) (12R-lipoxygenase) (12R-LOX) (Epidermis-type lipoxygenase 2) (e-LOX 2). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 18) | NC score | 0.120938 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
MCLN2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 19) | NC score | 0.278362 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 20) | NC score | 0.107034 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
MCLN2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 21) | NC score | 0.272470 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
RA6I1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.102268 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.097947 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.029246 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.100587 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
LIPE_MOUSE
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.048975 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVG5, Q8VDU2 | Gene names | Lipg | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL). | |||||
|
LOXE3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.148842 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BYJ1, Q9H4F2, Q9HC22 | Gene names | ALOXE3 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermis-type lipoxygenase 3 (EC 1.13.11.-) (e-LOX-3). | |||||
|
LOXE3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.156018 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WV07 | Gene names | Aloxe3 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermis-type lipoxygenase 3 (EC 1.13.11.-) (e-LOX-3). | |||||
|
CP3A7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.011952 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24462 | Gene names | CYP3A7 | |||
|
Domain Architecture |
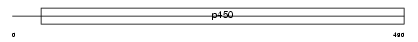
|
|||||
| Description | Cytochrome P450 3A7 (EC 1.14.14.1) (CYPIIIA7) (P450-HFLA). | |||||
|
LX15B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.146692 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35936 | Gene names | Alox15b, Alox8 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase type II (EC 1.13.11.33) (15-LOX-2) (8S- lipoxygenase) (8S-LOX). | |||||
|
LOX12_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.138467 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P18054, O95569, Q6ISF8, Q9UQM4 | Gene names | ALOX12, LOG12 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12S-type (EC 1.13.11.31) (12-LOX) (Platelet-type lipoxygenase 12). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.093349 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
TAP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.014702 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03518, Q16149 | Gene names | TAP1, ABCB2, PSF1, RING4, Y3 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen peptide transporter 1 (APT1) (Peptide transporter TAP1) (ATP- binding cassette sub-family B member 2) (Peptide transporter PSF1) (Peptide supply factor 1) (PSF-1) (Peptide transporter involved in antigen processing 1). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.093052 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
LIPE_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.048837 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5X9, Q6UW82 | Gene names | LIPG | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL) (EL). | |||||
|
NOS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.013994 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.093793 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.093240 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
DOS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.038840 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N350, O43385 | Gene names | DOS, C19orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dos. | |||||
|
EWS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.026129 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.025554 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
LX12L_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.136673 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P39654, Q4FJY9, Q5F2E3, Q6PHB2 | Gene names | Alox12l, Alox15 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, leukocyte-type (EC 1.13.11.31) (12-LOX). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.087595 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.087104 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.087215 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SDK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.005825 (rank : 85) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z5N4, Q8TEN9, Q8TEP5, Q96N44 | Gene names | SDK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.083504 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.083806 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
FBXL8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.026938 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96CD0, Q9NUM0 | Gene names | FBXL8, FBL8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 8 (F-box and leucine-rich repeat protein 8) (F-box protein FBL8). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.004104 (rank : 88) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
U689_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.034667 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D3J8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein UNQ689/PRO1329 homolog precursor. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.003015 (rank : 92) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
BAIP3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.014218 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80TT2, Q6RUT4 | Gene names | Baiap3, Gm937, Kiaa0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
LX15B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.142244 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15296, Q8IYQ2, Q8TEV3, Q8TEV4, Q8TEV5, Q8TEV6, Q9UKM4 | Gene names | ALOX15B | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase type II (EC 1.13.11.33) (15-LOX-2) (15- lipoxygenase 2). | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.007967 (rank : 83) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
LOX12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.134806 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P39655, Q8BHG4 | Gene names | Alox12, Alox12p | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12S-type (EC 1.13.11.31) (12-LOX) (Platelet-type lipoxygenase 12). | |||||
|
O51B2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | -0.000680 (rank : 94) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5P1, Q96RD4 | Gene names | OR51B2 | |||
|
Domain Architecture |
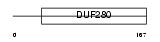
|
|||||
| Description | Olfactory receptor 51B2 (Odorant receptor HOR5'beta3). | |||||
|
OTU7A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.014683 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
CALR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.012648 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
DIP2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.018487 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14689, Q8IVA3, Q8N4S2, Q8TD89, Q96ML9 | Gene names | DIP2A, C21orf106, DIP2, KIAA0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
DIP2A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.018824 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BWT5, Q8CHI0 | Gene names | Dip2a, Dip2, Kiaa0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
NEP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.008204 (rank : 82) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61391, Q6NXX5, Q8K251 | Gene names | Mme | |||
|
Domain Architecture |

|
|||||
| Description | Neprilysin (EC 3.4.24.11) (Neutral endopeptidase) (NEP) (Enkephalinase) (Neutral endopeptidase 24.11) (Atriopeptidase) (CD10 antigen). | |||||
|
ABCG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.007565 (rank : 84) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64343 | Gene names | Abcg1, Abc8, Wht1 | |||
|
Domain Architecture |
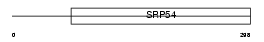
|
|||||
| Description | ATP-binding cassette sub-family G member 1 (White protein homolog) (ATP-binding cassette transporter 8). | |||||
|
PACS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.013355 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
PAIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.013074 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.094477 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.011737 (rank : 80) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ANDR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.003046 (rank : 91) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19091 | Gene names | Ar, Nr3c4 | |||
|
Domain Architecture |

|
|||||
| Description | Androgen receptor (Dihydrotestosterone receptor). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.072104 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
LAP4B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.011950 (rank : 79) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86VI4, Q3ZCV5, Q7L909, Q86VH8, Q9H060 | Gene names | LAPTM4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysosomal-associated transmembrane protein 4B (Lysosome-associated transmembrane protein 4-beta). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.004749 (rank : 87) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.009968 (rank : 81) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
NFASC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.002752 (rank : 93) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94856, Q5T2F0, Q5T2F1, Q5T2F2, Q5T2F3, Q5T2F4, Q5T2F5, Q5T2F6, Q5T2F7, Q5T2F9, Q5T2G0, Q5W9F8, Q68DH3, Q6ZQV6, Q7Z3K1, Q96HT1, Q96K50 | Gene names | NFASC, KIAA0756 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.003784 (rank : 90) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
TBX18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.003930 (rank : 89) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |
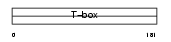
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.004977 (rank : 86) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.065200 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.065501 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.074000 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.070533 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.072087 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.078353 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.068896 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.072182 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.078926 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.061383 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.054337 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.069511 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.065576 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.065056 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
LOX15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.125575 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16050, Q8N6R7, Q99657 | Gene names | ALOX15, LOG15 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase (EC 1.13.11.33) (Arachidonate omega-6 lipoxygenase) (15-LOX). | |||||
|
LX12E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.129563 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P55249, Q91YW6 | Gene names | Alox12e, Alox12-ps2, Aloxe | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, epidermal-type (EC 1.13.11.31) (12-LOX). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.096677 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.070325 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.984787 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.586976 (rank : 3) | θ value | 3.07116e-24 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.579567 (rank : 4) | θ value | 2.43343e-21 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.577463 (rank : 5) | θ value | 9.24701e-21 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.574301 (rank : 6) | θ value | 6.40375e-22 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.571020 (rank : 7) | θ value | 2.97466e-19 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.559714 (rank : 8) | θ value | 5.60996e-18 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.324611 (rank : 9) | θ value | 1.92812e-18 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
MCLN1_HUMAN
|
||||||
| NC score | 0.303518 (rank : 10) | θ value | 9.92553e-07 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9GZU1, Q7Z4F7, Q9H292, Q9H4B3, Q9H4B5 | Gene names | MCOLN1, ML4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin) (MG-2). | |||||
|
MCLN1_MOUSE
|
||||||
| NC score | 0.296773 (rank : 11) | θ value | 4.92598e-06 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99J21 | Gene names | Mcoln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin). | |||||
|
MCLN3_HUMAN
|
||||||
| NC score | 0.296263 (rank : 12) | θ value | 0.000158464 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDD5, Q5T4H5, Q5T4H6, Q9NV09 | Gene names | MCOLN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN3_MOUSE
|
||||||
| NC score | 0.291543 (rank : 13) | θ value | 0.000270298 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN2_HUMAN
|
||||||
| NC score | 0.278362 (rank : 14) | θ value | 0.00298849 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
MCLN2_MOUSE
|
||||||
| NC score | 0.272470 (rank : 15) | θ value | 0.0252991 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
LOX5_HUMAN
|
||||||
| NC score | 0.250935 (rank : 16) | θ value | 1.74796e-11 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P09917 | Gene names | ALOX5, LOG5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
LOX5_MOUSE
|
||||||
| NC score | 0.240116 (rank : 17) | θ value | 4.30538e-10 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48999 | Gene names | Alox5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
LX12B_HUMAN
|
||||||
| NC score | 0.177979 (rank : 18) | θ value | 7.1131e-05 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75342 | Gene names | ALOX12B | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12R type (EC 1.13.11.-) (Epidermis-type lipoxygenase 12) (12R-lipoxygenase) (12R-LOX). | |||||
|
LX12B_MOUSE
|
||||||
| NC score | 0.170147 (rank : 19) | θ value | 0.00134147 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70582 | Gene names | Alox12b, Aloxe2 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12R type (EC 1.13.11.-) (Epidermis-type lipoxygenase 12) (12R-lipoxygenase) (12R-LOX) (Epidermis-type lipoxygenase 2) (e-LOX 2). | |||||
|
LOXE3_MOUSE
|
||||||
| NC score | 0.156018 (rank : 20) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WV07 | Gene names | Aloxe3 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermis-type lipoxygenase 3 (EC 1.13.11.-) (e-LOX-3). | |||||
|
LOXE3_HUMAN
|
||||||
| NC score | 0.148842 (rank : 21) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BYJ1, Q9H4F2, Q9HC22 | Gene names | ALOXE3 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermis-type lipoxygenase 3 (EC 1.13.11.-) (e-LOX-3). | |||||
|
LX15B_MOUSE
|
||||||
| NC score | 0.146692 (rank : 22) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35936 | Gene names | Alox15b, Alox8 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase type II (EC 1.13.11.33) (15-LOX-2) (8S- lipoxygenase) (8S-LOX). | |||||
|
LX15B_HUMAN
|
||||||
| NC score | 0.142244 (rank : 23) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15296, Q8IYQ2, Q8TEV3, Q8TEV4, Q8TEV5, Q8TEV6, Q9UKM4 | Gene names | ALOX15B | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase type II (EC 1.13.11.33) (15-LOX-2) (15- lipoxygenase 2). | |||||
|
LOX12_HUMAN
|
||||||
| NC score | 0.138467 (rank : 24) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P18054, O95569, Q6ISF8, Q9UQM4 | Gene names | ALOX12, LOG12 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12S-type (EC 1.13.11.31) (12-LOX) (Platelet-type lipoxygenase 12). | |||||
|
LX12L_MOUSE
|
||||||
| NC score | 0.136673 (rank : 25) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P39654, Q4FJY9, Q5F2E3, Q6PHB2 | Gene names | Alox12l, Alox15 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, leukocyte-type (EC 1.13.11.31) (12-LOX). | |||||
|
LOX12_MOUSE
|
||||||
| NC score | 0.134806 (rank : 26) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P39655, Q8BHG4 | Gene names | Alox12, Alox12p | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12S-type (EC 1.13.11.31) (12-LOX) (Platelet-type lipoxygenase 12). | |||||
|
LX12E_MOUSE
|
||||||
| NC score | 0.129563 (rank : 27) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P55249, Q91YW6 | Gene names | Alox12e, Alox12-ps2, Aloxe | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, epidermal-type (EC 1.13.11.31) (12-LOX). | |||||
|
LOX15_HUMAN
|
||||||
| NC score | 0.125575 (rank : 28) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16050, Q8N6R7, Q99657 | Gene names | ALOX15, LOG15 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase (EC 1.13.11.33) (Arachidonate omega-6 lipoxygenase) (15-LOX). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.120938 (rank : 29) | θ value | 0.00175202 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.107034 (rank : 30) | θ value | 0.0148317 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
RA6I1_HUMAN
|
||||||
| NC score | 0.102268 (rank : 31) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.100587 (rank : 32) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.097947 (rank : 33) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.096677 (rank : 34) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.094477 (rank : 35) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.093793 (rank : 36) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.093349 (rank : 37) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.093240 (rank : 38) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.093052 (rank : 39) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.087595 (rank : 40) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.087215 (rank : 41) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.087104 (rank : 42) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.083806 (rank : 43) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.083504 (rank : 44) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.078926 (rank : 45) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.078353 (rank : 46) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.074000 (rank : 47) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.072182 (rank : 48) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.072104 (rank : 49) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.072087 (rank : 50) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.070533 (rank : 51) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.070325 (rank : 52) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.069511 (rank : 53) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.068896 (rank : 54) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.065576 (rank : 55) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.065501 (rank : 56) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.065200 (rank : 57) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.065056 (rank : 58) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.061383 (rank : 59) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.054337 (rank : 60) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
LIPE_MOUSE
|
||||||
| NC score | 0.048975 (rank : 61) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVG5, Q8VDU2 | Gene names | Lipg | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL). | |||||
|
LIPE_HUMAN
|
||||||
| NC score | 0.048837 (rank : 62) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5X9, Q6UW82 | Gene names | LIPG | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL) (EL). | |||||
|
DOS_HUMAN
|
||||||
| NC score | 0.038840 (rank : 63) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N350, O43385 | Gene names | DOS, C19orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dos. | |||||
|
U689_MOUSE
|
||||||
| NC score | 0.034667 (rank : 64) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D3J8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein UNQ689/PRO1329 homolog precursor. | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.029246 (rank : 65) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
FBXL8_HUMAN
|
||||||
| NC score | 0.026938 (rank : 66) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96CD0, Q9NUM0 | Gene names | FBXL8, FBL8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 8 (F-box and leucine-rich repeat protein 8) (F-box protein FBL8). | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.026129 (rank : 67) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.025554 (rank : 68) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
DIP2A_MOUSE
|
||||||
| NC score | 0.018824 (rank : 69) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BWT5, Q8CHI0 | Gene names | Dip2a, Dip2, Kiaa0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
DIP2A_HUMAN
|
||||||
| NC score | 0.018487 (rank : 70) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14689, Q8IVA3, Q8N4S2, Q8TD89, Q96ML9 | Gene names | DIP2A, C21orf106, DIP2, KIAA0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
TAP1_HUMAN
|
||||||
| NC score | 0.014702 (rank : 71) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03518, Q16149 | Gene names | TAP1, ABCB2, PSF1, RING4, Y3 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen peptide transporter 1 (APT1) (Peptide transporter TAP1) (ATP- binding cassette sub-family B member 2) (Peptide transporter PSF1) (Peptide supply factor 1) (PSF-1) (Peptide transporter involved in antigen processing 1). | |||||
|
OTU7A_MOUSE
|
||||||
| NC score | 0.014683 (rank : 72) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
BAIP3_MOUSE
|
||||||
| NC score | 0.014218 (rank : 73) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80TT2, Q6RUT4 | Gene names | Baiap3, Gm937, Kiaa0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
NOS1_HUMAN
|
||||||
| NC score | 0.013994 (rank : 74) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
PACS1_MOUSE
|
||||||
| NC score | 0.013355 (rank : 75) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
PAIP1_HUMAN
|
||||||
| NC score | 0.013074 (rank : 76) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
CALR3_MOUSE
|
||||||
| NC score | 0.012648 (rank : 77) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CP3A7_HUMAN
|
||||||
| NC score | 0.011952 (rank : 78) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24462 | Gene names | CYP3A7 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A7 (EC 1.14.14.1) (CYPIIIA7) (P450-HFLA). | |||||
|
LAP4B_HUMAN
|
||||||
| NC score | 0.011950 (rank : 79) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86VI4, Q3ZCV5, Q7L909, Q86VH8, Q9H060 | Gene names | LAPTM4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysosomal-associated transmembrane protein 4B (Lysosome-associated transmembrane protein 4-beta). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.011737 (rank : 80) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.009968 (rank : 81) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
NEP_MOUSE
|
||||||
| NC score | 0.008204 (rank : 82) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61391, Q6NXX5, Q8K251 | Gene names | Mme | |||
|
Domain Architecture |

|
|||||
| Description | Neprilysin (EC 3.4.24.11) (Neutral endopeptidase) (NEP) (Enkephalinase) (Neutral endopeptidase 24.11) (Atriopeptidase) (CD10 antigen). | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.007967 (rank : 83) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
ABCG1_MOUSE
|
||||||
| NC score | 0.007565 (rank : 84) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64343 | Gene names | Abcg1, Abc8, Wht1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family G member 1 (White protein homolog) (ATP-binding cassette transporter 8). | |||||
|
SDK1_HUMAN
|
||||||
| NC score | 0.005825 (rank : 85) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z5N4, Q8TEN9, Q8TEP5, Q96N44 | Gene names | SDK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.004977 (rank : 86) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.004749 (rank : 87) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.004104 (rank : 88) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
TBX18_HUMAN
|
||||||
| NC score | 0.003930 (rank : 89) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.003784 (rank : 90) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
ANDR_MOUSE
|
||||||
| NC score | 0.003046 (rank : 91) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19091 | Gene names | Ar, Nr3c4 | |||
|
Domain Architecture |

|
|||||
| Description | Androgen receptor (Dihydrotestosterone receptor). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.003015 (rank : 92) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
NFASC_HUMAN
|
||||||
| NC score | 0.002752 (rank : 93) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94856, Q5T2F0, Q5T2F1, Q5T2F2, Q5T2F3, Q5T2F4, Q5T2F5, Q5T2F6, Q5T2F7, Q5T2F9, Q5T2G0, Q5W9F8, Q68DH3, Q6ZQV6, Q7Z3K1, Q96HT1, Q96K50 | Gene names | NFASC, KIAA0756 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
O51B2_HUMAN
|
||||||
| NC score | -0.000680 (rank : 94) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5P1, Q96RD4 | Gene names | OR51B2 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 51B2 (Odorant receptor HOR5'beta3). | |||||