Please be patient as the page loads
|
PK2L1_HUMAN
|
||||||
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PK2L1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.969112 (rank : 2) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.967132 (rank : 3) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 1.42583e-138 (rank : 4) | NC score | 0.961548 (rank : 5) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 3.4016e-132 (rank : 5) | NC score | 0.963751 (rank : 4) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 6) | NC score | 0.601621 (rank : 6) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 7) | NC score | 0.574301 (rank : 7) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 8) | NC score | 0.315724 (rank : 27) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 9) | NC score | 0.470979 (rank : 8) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 10) | NC score | 0.318730 (rank : 26) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
TRPC3_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 11) | NC score | 0.252901 (rank : 47) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 12) | NC score | 0.357536 (rank : 12) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 13) | NC score | 0.358209 (rank : 11) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
MCLN2_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 14) | NC score | 0.360002 (rank : 10) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
MCLN2_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 15) | NC score | 0.352623 (rank : 16) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
TRPC3_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 16) | NC score | 0.254861 (rank : 46) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 17) | NC score | 0.355677 (rank : 13) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
MCLN3_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 18) | NC score | 0.346492 (rank : 20) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 19) | NC score | 0.361301 (rank : 9) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 20) | NC score | 0.342728 (rank : 21) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 21) | NC score | 0.354260 (rank : 15) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 22) | NC score | 0.355071 (rank : 14) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 23) | NC score | 0.312251 (rank : 29) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 24) | NC score | 0.340281 (rank : 23) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
MCLN3_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 25) | NC score | 0.347862 (rank : 18) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDD5, Q5T4H5, Q5T4H6, Q9NV09 | Gene names | MCOLN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 26) | NC score | 0.351437 (rank : 17) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 27) | NC score | 0.342641 (rank : 22) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
TRPC7_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 28) | NC score | 0.220531 (rank : 53) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC7_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 29) | NC score | 0.220906 (rank : 52) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |
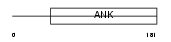
|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 30) | NC score | 0.347683 (rank : 19) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 31) | NC score | 0.327045 (rank : 25) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 32) | NC score | 0.329498 (rank : 24) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
TRPA1_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 33) | NC score | 0.121080 (rank : 71) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
TRPC4_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 34) | NC score | 0.238027 (rank : 48) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 35) | NC score | 0.237641 (rank : 49) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC6_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 36) | NC score | 0.233716 (rank : 51) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 37) | NC score | 0.234518 (rank : 50) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
MCLN1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 38) | NC score | 0.312899 (rank : 28) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9GZU1, Q7Z4F7, Q9H292, Q9H4B3, Q9H4B5 | Gene names | MCOLN1, ML4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin) (MG-2). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 39) | NC score | 0.309196 (rank : 30) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 40) | NC score | 0.138072 (rank : 66) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 41) | NC score | 0.141675 (rank : 64) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
MCLN1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 42) | NC score | 0.308039 (rank : 31) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99J21 | Gene names | Mcoln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin). | |||||
|
TRPV5_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 43) | NC score | 0.168057 (rank : 58) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPA1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 44) | NC score | 0.113459 (rank : 72) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 45) | NC score | 0.282681 (rank : 37) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 46) | NC score | 0.282374 (rank : 38) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
TRPV6_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 47) | NC score | 0.132505 (rank : 67) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 48) | NC score | 0.285318 (rank : 34) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 49) | NC score | 0.280660 (rank : 40) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 50) | NC score | 0.182617 (rank : 56) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 51) | NC score | 0.140644 (rank : 65) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV6_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 52) | NC score | 0.123338 (rank : 70) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 53) | NC score | 0.281953 (rank : 39) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 54) | NC score | 0.283570 (rank : 36) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 55) | NC score | 0.167036 (rank : 59) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 56) | NC score | 0.292135 (rank : 32) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 57) | NC score | 0.273156 (rank : 42) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 58) | NC score | 0.272706 (rank : 43) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 59) | NC score | 0.274069 (rank : 41) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 60) | NC score | 0.152224 (rank : 62) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 61) | NC score | 0.151087 (rank : 63) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 62) | NC score | 0.267650 (rank : 45) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 0.21417 (rank : 63) | NC score | 0.269835 (rank : 44) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 0.279714 (rank : 64) | NC score | 0.034438 (rank : 86) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.030429 (rank : 91) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 66) | NC score | 0.286213 (rank : 33) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
TRPC5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 67) | NC score | 0.202660 (rank : 54) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 68) | NC score | 0.202543 (rank : 55) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
CD2AP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 69) | NC score | 0.016246 (rank : 125) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5K6, Q9UG97 | Gene names | CD2AP | |||
|
Domain Architecture |
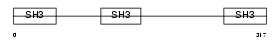
|
|||||
| Description | CD2-associated protein (Cas ligand with multiple SH3 domains) (Adapter protein CMS). | |||||
|
BICD2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 70) | NC score | 0.035606 (rank : 84) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 71) | NC score | 0.035856 (rank : 83) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
TRPM8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.124972 (rank : 68) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
OFD1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 73) | NC score | 0.035235 (rank : 85) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 74) | NC score | 0.284783 (rank : 35) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.025650 (rank : 102) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TRPM8_HUMAN
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.124195 (rank : 69) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
REST_MOUSE
|
||||||
| θ value | 0.813845 (rank : 77) | NC score | 0.026235 (rank : 98) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.025030 (rank : 103) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.031295 (rank : 89) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 80) | NC score | 0.025749 (rank : 101) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.023827 (rank : 106) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.033620 (rank : 87) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.023816 (rank : 107) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.023640 (rank : 108) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | -0.000512 (rank : 143) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
CCD39_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.028777 (rank : 93) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.026014 (rank : 100) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
LETM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.030054 (rank : 92) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95202, Q9UF65 | Gene names | LETM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
NIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.030531 (rank : 90) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.015380 (rank : 126) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
CALB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.050738 (rank : 82) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.026554 (rank : 97) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.011911 (rank : 127) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.023281 (rank : 109) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
SNX23_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.016514 (rank : 123) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.010698 (rank : 129) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
MRP9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.010590 (rank : 130) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J65, Q49AL2, Q8TAF0, Q8TEY2 | Gene names | ABCC12, MRP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multidrug resistance-associated protein 9 (ATP-binding cassette transporter sub-family C member 12). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.022755 (rank : 111) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.019434 (rank : 121) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.016460 (rank : 124) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.024635 (rank : 104) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.026129 (rank : 99) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.010384 (rank : 131) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.022609 (rank : 112) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.019854 (rank : 120) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.027827 (rank : 95) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.000296 (rank : 142) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
FA50A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.022403 (rank : 113) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14320, Q5HY37 | Gene names | FAM50A, DXS9928E, HXC26, XAP5 | |||
|
Domain Architecture |
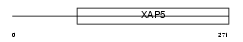
|
|||||
| Description | Protein FAM50A (Protein XAP-5) (Protein HXC-26). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.022287 (rank : 114) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.020420 (rank : 119) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.033549 (rank : 88) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.006433 (rank : 137) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
SNX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.009187 (rank : 134) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WV80, Q9EQZ9 | Gene names | Snx1 | |||
|
Domain Architecture |
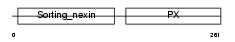
|
|||||
| Description | Sorting nexin-1. | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.002407 (rank : 141) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.022893 (rank : 110) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
IRX3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.004644 (rank : 139) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
LAMB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.006676 (rank : 136) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.021383 (rank : 117) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.026884 (rank : 96) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.018308 (rank : 122) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
TRPM3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.091896 (rank : 78) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TTP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.009563 (rank : 133) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
ARY3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.009681 (rank : 132) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50296 | Gene names | Nat3, Aac3 | |||
|
Domain Architecture |

|
|||||
| Description | Arylamine N-acetyltransferase 3 (EC 2.3.1.5) (Arylamide acetylase 3) (N-acetyltransferase type 3) (NAT-3). | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.005369 (rank : 138) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.003463 (rank : 140) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.021395 (rank : 116) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.021832 (rank : 115) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NARGL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.008236 (rank : 135) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6N069, Q5VSP9, Q6P2D5, Q8N5J3, Q8N870 | Gene names | NARG1L, NAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.027877 (rank : 94) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.021156 (rank : 118) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.011877 (rank : 128) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.024115 (rank : 105) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.159445 (rank : 61) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.159548 (rank : 60) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.168088 (rank : 57) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.093189 (rank : 76) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.091852 (rank : 79) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.092033 (rank : 77) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.093806 (rank : 75) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPV1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.099083 (rank : 73) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.094341 (rank : 74) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPV2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.080817 (rank : 81) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.081141 (rank : 80) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.969112 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.967132 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.963751 (rank : 4) | θ value | 3.4016e-132 (rank : 5) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.961548 (rank : 5) | θ value | 1.42583e-138 (rank : 4) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.601621 (rank : 6) | θ value | 2.12685e-25 (rank : 6) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.574301 (rank : 7) | θ value | 6.40375e-22 (rank : 7) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.470979 (rank : 8) | θ value | 1.63604e-09 (rank : 9) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.361301 (rank : 9) | θ value | 4.92598e-06 (rank : 19) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
MCLN2_MOUSE
|
||||||
| NC score | 0.360002 (rank : 10) | θ value | 9.92553e-07 (rank : 14) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.358209 (rank : 11) | θ value | 9.92553e-07 (rank : 13) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.357536 (rank : 12) | θ value | 9.92553e-07 (rank : 12) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.355677 (rank : 13) | θ value | 2.88788e-06 (rank : 17) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.355071 (rank : 14) | θ value | 1.87187e-05 (rank : 22) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.354260 (rank : 15) | θ value | 1.43324e-05 (rank : 21) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
MCLN2_HUMAN
|
||||||
| NC score | 0.352623 (rank : 16) | θ value | 1.29631e-06 (rank : 15) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.351437 (rank : 17) | θ value | 5.44631e-05 (rank : 26) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
MCLN3_HUMAN
|
||||||
| NC score | 0.347862 (rank : 18) | θ value | 3.19293e-05 (rank : 25) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDD5, Q5T4H5, Q5T4H6, Q9NV09 | Gene names | MCOLN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.347683 (rank : 19) | θ value | 9.29e-05 (rank : 30) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
MCLN3_MOUSE
|
||||||
| NC score | 0.346492 (rank : 20) | θ value | 3.77169e-06 (rank : 18) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.342728 (rank : 21) | θ value | 4.92598e-06 (rank : 20) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.342641 (rank : 22) | θ value | 5.44631e-05 (rank : 27) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.340281 (rank : 23) | θ value | 3.19293e-05 (rank : 24) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.329498 (rank : 24) | θ value | 0.000158464 (rank : 32) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.327045 (rank : 25) | θ value | 0.000158464 (rank : 31) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.318730 (rank : 26) | θ value | 1.99992e-07 (rank : 10) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.315724 (rank : 27) | θ value | 2.7842e-17 (rank : 8) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
MCLN1_HUMAN
|
||||||
| NC score | 0.312899 (rank : 28) | θ value | 0.00035302 (rank : 38) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9GZU1, Q7Z4F7, Q9H292, Q9H4B3, Q9H4B5 | Gene names | MCOLN1, ML4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin) (MG-2). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.312251 (rank : 29) | θ value | 3.19293e-05 (rank : 23) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.309196 (rank : 30) | θ value | 0.00035302 (rank : 39) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
MCLN1_MOUSE
|
||||||
| NC score | 0.308039 (rank : 31) | θ value | 0.00102713 (rank : 42) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99J21 | Gene names | Mcoln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.292135 (rank : 32) | θ value | 0.0563607 (rank : 56) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.286213 (rank : 33) | θ value | 0.279714 (rank : 66) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.285318 (rank : 34) | θ value | 0.0113563 (rank : 48) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.284783 (rank : 35) | θ value | 0.62314 (rank : 74) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.283570 (rank : 36) | θ value | 0.0431538 (rank : 54) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.282681 (rank : 37) | θ value | 0.00665767 (rank : 45) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.282374 (rank : 38) | θ value | 0.00665767 (rank : 46) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.281953 (rank : 39) | θ value | 0.0330416 (rank : 53) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.280660 (rank : 40) | θ value | 0.0193708 (rank : 49) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.274069 (rank : 41) | θ value | 0.0961366 (rank : 59) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.273156 (rank : 42) | θ value | 0.0736092 (rank : 57) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.272706 (rank : 43) | θ value | 0.0736092 (rank : 58) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.269835 (rank : 44) | θ value | 0.21417 (rank : 63) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.267650 (rank : 45) | θ value | 0.163984 (rank : 62) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
TRPC3_MOUSE
|
||||||
| NC score | 0.254861 (rank : 46) | θ value | 1.69304e-06 (rank : 16) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC3_HUMAN
|
||||||
| NC score | 0.252901 (rank : 47) | θ value | 7.59969e-07 (rank : 11) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.238027 (rank : 48) | θ value | 0.000270298 (rank : 34) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.237641 (rank : 49) | θ value | 0.000270298 (rank : 35) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.234518 (rank : 50) | θ value | 0.000270298 (rank : 37) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC6_HUMAN
|
||||||
| NC score | 0.233716 (rank : 51) | θ value | 0.000270298 (rank : 36) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC7_MOUSE
|
||||||
| NC score | 0.220906 (rank : 52) | θ value | 7.1131e-05 (rank : 29) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC7_HUMAN
|
||||||
| NC score | 0.220531 (rank : 53) | θ value | 7.1131e-05 (rank : 28) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC5_HUMAN
|
||||||
| NC score | 0.202660 (rank : 54) | θ value | 0.279714 (rank : 67) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC5_MOUSE
|
||||||
| NC score | 0.202543 (rank : 55) | θ value | 0.279714 (rank : 68) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.182617 (rank : 56) | θ value | 0.0193708 (rank : 50) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPC2_MOUSE
|
||||||
| NC score | 0.168088 (rank : 57) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPV5_MOUSE
|
||||||
| NC score | 0.168057 (rank : 58) | θ value | 0.00134147 (rank : 43) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.167036 (rank : 59) | θ value | 0.0431538 (rank : 55) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.159548 (rank : 60) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.159445 (rank : 61) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.152224 (rank : 62) | θ value | 0.0961366 (rank : 60) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.151087 (rank : 63) | θ value | 0.125558 (rank : 61) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.141675 (rank : 64) | θ value | 0.000786445 (rank : 41) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.140644 (rank : 65) | θ value | 0.0193708 (rank : 51) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.138072 (rank : 66) | θ value | 0.000786445 (rank : 40) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPV6_HUMAN
|
||||||
| NC score | 0.132505 (rank : 67) | θ value | 0.00869519 (rank : 47) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPM8_MOUSE
|
||||||
| NC score | 0.124972 (rank : 68) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPM8_HUMAN
|
||||||
| NC score | 0.124195 (rank : 69) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPV6_MOUSE
|
||||||
| NC score | 0.123338 (rank : 70) | θ value | 0.0252991 (rank : 52) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
TRPA1_HUMAN
|
||||||
| NC score | 0.121080 (rank : 71) | θ value | 0.000158464 (rank : 33) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
TRPA1_MOUSE
|
||||||
| NC score | 0.113459 (rank : 72) | θ value | 0.00228821 (rank : 44) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
TRPV1_HUMAN
|
||||||
| NC score | 0.099083 (rank : 73) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV1_MOUSE
|
||||||
| NC score | 0.094341 (rank : 74) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPM6_MOUSE
|
||||||
| NC score | 0.093806 (rank : 75) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM2_HUMAN
|
||||||
| NC score | 0.093189 (rank : 76) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM6_HUMAN
|
||||||
| NC score | 0.092033 (rank : 77) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM3_HUMAN
|
||||||
| NC score | 0.091896 (rank : 78) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPM2_MOUSE
|
||||||
| NC score | 0.091852 (rank : 79) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.081141 (rank : 80) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
TRPV2_HUMAN
|
||||||
| NC score | 0.080817 (rank : 81) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.050738 (rank : 82) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.035856 (rank : 83) | θ value | 0.47712 (rank : 71) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
BICD2_HUMAN
|
||||||
| NC score | 0.035606 (rank : 84) | θ value | 0.47712 (rank : 70) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.035235 (rank : 85) | θ value | 0.62314 (rank : 73) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.034438 (rank : 86) | θ value | 0.279714 (rank : 64) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.033620 (rank : 87) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.033549 (rank : 88) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.031295 (rank : 89) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.030531 (rank : 90) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.030429 (rank : 91) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
LETM1_HUMAN
|
||||||
| NC score | 0.030054 (rank : 92) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95202, Q9UF65 | Gene names | LETM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.028777 (rank : 93) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.027877 (rank : 94) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.027827 (rank : 95) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.026884 (rank : 96) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.026554 (rank : 97) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.026235 (rank : 98) | θ value | 0.813845 (rank : 77) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.026129 (rank : 99) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.026014 (rank : 100) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.025749 (rank : 101) | θ value | 1.06291 (rank : 80) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.025650 (rank : 102) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.025030 (rank : 103) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.024635 (rank : 104) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.024115 (rank : 105) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.023827 (rank : 106) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.023816 (rank : 107) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.023640 (rank : 108) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.023281 (rank : 109) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.022893 (rank : 110) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.022755 (rank : 111) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.022609 (rank : 112) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
FA50A_HUMAN
|
||||||
| NC score | 0.022403 (rank : 113) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14320, Q5HY37 | Gene names | FAM50A, DXS9928E, HXC26, XAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM50A (Protein XAP-5) (Protein HXC-26). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.022287 (rank : 114) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.021832 (rank : 115) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.021395 (rank : 116) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.021383 (rank : 117) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.021156 (rank : 118) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.020420 (rank : 119) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.019854 (rank : 120) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.019434 (rank : 121) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.018308 (rank : 122) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
SNX23_HUMAN
|
||||||
| NC score | 0.016514 (rank : 123) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.016460 (rank : 124) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CD2AP_HUMAN
|
||||||
| NC score | 0.016246 (rank : 125) | θ value | 0.365318 (rank : 69) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5K6, Q9UG97 | Gene names | CD2AP | |||
|
Domain Architecture |

|
|||||
| Description | CD2-associated protein (Cas ligand with multiple SH3 domains) (Adapter protein CMS). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.015380 (rank : 126) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.011911 (rank : 127) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.011877 (rank : 128) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.010698 (rank : 129) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
MRP9_HUMAN
|
||||||
| NC score | 0.010590 (rank : 130) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J65, Q49AL2, Q8TAF0, Q8TEY2 | Gene names | ABCC12, MRP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multidrug resistance-associated protein 9 (ATP-binding cassette transporter sub-family C member 12). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.010384 (rank : 131) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
ARY3_MOUSE
|
||||||
| NC score | 0.009681 (rank : 132) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50296 | Gene names | Nat3, Aac3 | |||
|
Domain Architecture |

|
|||||
| Description | Arylamine N-acetyltransferase 3 (EC 2.3.1.5) (Arylamide acetylase 3) (N-acetyltransferase type 3) (NAT-3). | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.009563 (rank : 133) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
SNX1_MOUSE
|
||||||
| NC score | 0.009187 (rank : 134) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WV80, Q9EQZ9 | Gene names | Snx1 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-1. | |||||
|
NARGL_HUMAN
|
||||||
| NC score | 0.008236 (rank : 135) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6N069, Q5VSP9, Q6P2D5, Q8N5J3, Q8N870 | Gene names | NARG1L, NAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
LAMB3_HUMAN
|
||||||
| NC score | 0.006676 (rank : 136) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.006433 (rank : 137) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.005369 (rank : 138) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
IRX3_MOUSE
|
||||||
| NC score | 0.004644 (rank : 139) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.003463 (rank : 140) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.002407 (rank : 141) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.000296 (rank : 142) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | -0.000512 (rank : 143) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||