Please be patient as the page loads
|
SCN1A_HUMAN
|
||||||
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SC10A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992373 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991929 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.983180 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.984738 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.997105 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.996482 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.991367 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.986532 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.985313 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.994527 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.995339 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 0 (rank : 13) | NC score | 0.994925 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 4.99229e-75 (rank : 14) | NC score | 0.851868 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 1.11217e-74 (rank : 15) | NC score | 0.844102 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 3.57772e-73 (rank : 16) | NC score | 0.893084 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 1.96316e-71 (rank : 17) | NC score | 0.843867 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 1.66191e-70 (rank : 18) | NC score | 0.841975 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 1.66191e-70 (rank : 19) | NC score | 0.872736 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 3.70236e-70 (rank : 20) | NC score | 0.849262 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 3.13423e-69 (rank : 21) | NC score | 0.855226 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 3.13423e-69 (rank : 22) | NC score | 0.865624 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 4.09345e-69 (rank : 23) | NC score | 0.855745 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 9.11921e-69 (rank : 24) | NC score | 0.844435 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 6.53529e-67 (rank : 25) | NC score | 0.843179 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 1.11475e-66 (rank : 26) | NC score | 0.844847 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 1.36269e-64 (rank : 27) | NC score | 0.836675 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 1.50663e-63 (rank : 28) | NC score | 0.837717 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 2.56992e-63 (rank : 29) | NC score | 0.838548 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 5.92465e-60 (rank : 30) | NC score | 0.831669 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 31) | NC score | 0.209533 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 32) | NC score | 0.224962 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 33) | NC score | 0.189575 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 34) | NC score | 0.189668 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 35) | NC score | 0.170625 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 36) | NC score | 0.170740 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 37) | NC score | 0.282681 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 38) | NC score | 0.262004 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 39) | NC score | 0.031866 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 40) | NC score | 0.208234 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 41) | NC score | 0.304278 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 42) | NC score | 0.269667 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 43) | NC score | 0.199956 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.070510 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 45) | NC score | 0.070580 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 46) | NC score | 0.136618 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 47) | NC score | 0.018526 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 48) | NC score | 0.134289 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 49) | NC score | 0.163081 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 50) | NC score | 0.163247 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 51) | NC score | 0.194669 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 52) | NC score | 0.168405 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 53) | NC score | 0.028676 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
IMMT_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 54) | NC score | 0.025932 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16891, Q14539, Q15092, Q68D41, Q69HW5, Q6IBL0, Q7Z3X1, Q8TAJ5, Q9P0V2 | Gene names | IMMT, HMP, PIG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin) (p87/89) (Proliferation-inducing gene 4 protein). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 55) | NC score | 0.010016 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 56) | NC score | 0.019823 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 57) | NC score | 0.265334 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 58) | NC score | 0.058212 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 59) | NC score | 0.144689 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
MARK2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 60) | NC score | -0.002525 (rank : 166) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 61) | NC score | 0.092997 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
LAMC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 62) | NC score | 0.006295 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 63) | NC score | 0.142930 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | 0.279714 (rank : 64) | NC score | 0.119512 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
TPTE2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.139556 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
GLE1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 66) | NC score | 0.017782 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 67) | NC score | 0.042478 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.017137 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.011151 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
FA61A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.012788 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8ND56, Q76LX7, Q96AR3, Q96K73, Q96SN5, Q9UFR3 | Gene names | FAM61A, C19orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A (Putative alpha synuclein-binding protein) (AlphaSNBP). | |||||
|
TEX9_MOUSE
|
||||||
| θ value | 0.813845 (rank : 71) | NC score | 0.015784 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.064453 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.019847 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.132482 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.012812 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.005189 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.093793 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.005076 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.012962 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.006196 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.059692 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.061166 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.012291 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
A16L1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.011626 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C0J2, Q6KAT7, Q80U97, Q80U98, Q80U99, Q80Y53, Q9DB63 | Gene names | Atg16l1, Apg16l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.007564 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
GALR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | -0.003647 (rank : 168) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47211 | Gene names | GALR1, GALNR, GALNR1 | |||
|
Domain Architecture |
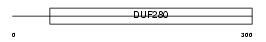
|
|||||
| Description | Galanin receptor type 1 (GAL1-R) (GALR1). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.013542 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
AR13B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.008457 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.011844 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.012497 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
LC7L2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.018063 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.010528 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.023028 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.027969 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PSN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.033352 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61144, O54977, P97934, P97935, Q91VS3, Q9D616 | Gene names | Psen2, Ad4h, Alg3, Ps-2, Psnl2 | |||
|
Domain Architecture |
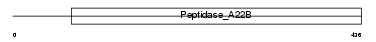
|
|||||
| Description | Presenilin-2 (EC 3.4.23.-) (PS-2) [Contains: Presenilin-2 NTF subunit; Presenilin-2 CTF subunit]. | |||||
|
TPTE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.095318 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
GP73_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.008392 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
LC7L2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.018661 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | -0.003537 (rank : 167) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.007260 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
SACS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.006306 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.009938 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.001822 (rank : 163) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.010172 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
BFAR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.008257 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZS9 | Gene names | BFAR, BAR, RNF47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator (RING finger protein 47). | |||||
|
BFAR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.008231 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.008594 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.007857 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.011959 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.008440 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.117006 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.110323 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.065355 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
MCA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.010422 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12904, Q6FG28, Q96CQ9 | Gene names | SCYE1, EMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.010018 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.006381 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.006288 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.002135 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TMCC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.006725 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULS5, Q8IWB2 | Gene names | TMCC3, KIAA1145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 3. | |||||
|
CN145_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.008232 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.012687 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.109281 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
MCA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.008452 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P31230, Q60659 | Gene names | Scye1, Emap2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.005875 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.020936 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.008748 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SP17_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.045655 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62252 | Gene names | Spa17, Sp17 | |||
|
Domain Architecture |
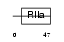
|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17). | |||||
|
A16L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.006517 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q676U5, Q6IPN1, Q6UXW4, Q6ZVZ5, Q8NCY2, Q96JV5, Q9H619 | Gene names | ATG16L1, APG16L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
ARD1H_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.004971 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41227 | Gene names | ARD1A, ARD1, TE2 | |||
|
Domain Architecture |
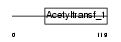
|
|||||
| Description | N-terminal acetyltransferase complex ARD1 subunit homolog A (EC 2.3.1.88) (EC 2.3.1.-). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.019225 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.019650 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.012113 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.005858 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.010017 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.004986 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.107204 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
LHX3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | -0.002085 (rank : 165) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |
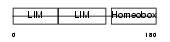
|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
MARK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | -0.003793 (rank : 169) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.004327 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
NELFE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.006308 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19426, P97485, P97486, Q9CZN3 | Gene names | Rdbp, D17h6s45, Nelfe, Rd | |||
|
Domain Architecture |

|
|||||
| Description | Negative elongation factor E (NELF-E) (RD protein). | |||||
|
NELF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.011547 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6X4W1, Q6X4V7, Q6X4V8, Q6X4V9, Q8N2M2, Q96SY1, Q9NPM4, Q9NPP3, Q9NPS3 | Gene names | NELF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor). | |||||
|
NELF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.013172 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99NF2, Q8BPT0, Q8C9R5, Q9DBF4, Q9ERZ1 | Gene names | Nelf, Jac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor) (Juxtasynaptic attractor of caldendrin on dendritic boutons protein) (Jacob protein). | |||||
|
NPC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.002736 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15118, Q9P130 | Gene names | NPC1 | |||
|
Domain Architecture |

|
|||||
| Description | Niemann-Pick C1 protein precursor. | |||||
|
OLF9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | -0.001729 (rank : 164) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60885, Q8VGC0 | Gene names | Olfr9, Mor269-3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 9 (Olfactory receptor 269-3) (Odorant receptor M25). | |||||
|
PML_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.002362 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29590, P29591, P29592, P29593, Q00755 | Gene names | PML, MYL, RNF71, TRIM19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.004257 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.003911 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ZWINT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.006577 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95229, Q9BWD0 | Gene names | ZWINT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.073353 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.074708 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.071452 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.072270 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.064433 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.063593 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.051130 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.051515 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.061050 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.065848 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.066469 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.069219 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.058675 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.085156 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.085543 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.057290 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.056032 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.064273 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.072293 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.050356 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.114053 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.997105 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.996482 (rank : 3) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.995339 (rank : 4) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.994925 (rank : 5) | θ value | 0 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.994527 (rank : 6) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.992373 (rank : 7) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.991929 (rank : 8) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.991367 (rank : 9) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.986532 (rank : 10) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.985313 (rank : 11) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.984738 (rank : 12) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.983180 (rank : 13) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.893084 (rank : 14) | θ value | 3.57772e-73 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.872736 (rank : 15) | θ value | 1.66191e-70 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.865624 (rank : 16) | θ value | 3.13423e-69 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.855745 (rank : 17) | θ value | 4.09345e-69 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.855226 (rank : 18) | θ value | 3.13423e-69 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.851868 (rank : 19) | θ value | 4.99229e-75 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.849262 (rank : 20) | θ value | 3.70236e-70 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.844847 (rank : 21) | θ value | 1.11475e-66 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.844435 (rank : 22) | θ value | 9.11921e-69 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.844102 (rank : 23) | θ value | 1.11217e-74 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.843867 (rank : 24) | θ value | 1.96316e-71 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.843179 (rank : 25) | θ value | 6.53529e-67 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.841975 (rank : 26) | θ value | 1.66191e-70 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.838548 (rank : 27) | θ value | 2.56992e-63 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.837717 (rank : 28) | θ value | 1.50663e-63 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.836675 (rank : 29) | θ value | 1.36269e-64 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.831669 (rank : 30) | θ value | 5.92465e-60 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.304278 (rank : 31) | θ value | 0.0113563 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.282681 (rank : 32) | θ value | 0.00665767 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.269667 (rank : 33) | θ value | 0.0113563 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.265334 (rank : 34) | θ value | 0.125558 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.262004 (rank : 35) | θ value | 0.00665767 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.224962 (rank : 36) | θ value | 7.1131e-05 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.209533 (rank : 37) | θ value | 1.09739e-05 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.208234 (rank : 38) | θ value | 0.0113563 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.199956 (rank : 39) | θ value | 0.0148317 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.194669 (rank : 40) | θ value | 0.0252991 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.189668 (rank : 41) | θ value | 0.00035302 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.189575 (rank : 42) | θ value | 0.00035302 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.170740 (rank : 43) | θ value | 0.00509761 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.170625 (rank : 44) | θ value | 0.00509761 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.168405 (rank : 45) | θ value | 0.0252991 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.163247 (rank : 46) | θ value | 0.0252991 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.163081 (rank : 47) | θ value | 0.0252991 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.144689 (rank : 48) | θ value | 0.163984 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.142930 (rank : 49) | θ value | 0.279714 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
TPTE2_HUMAN
|
||||||
| NC score | 0.139556 (rank : 50) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.136618 (rank : 51) | θ value | 0.0193708 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.134289 (rank : 52) | θ value | 0.0252991 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.132482 (rank : 53) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.119512 (rank : 54) | θ value | 0.279714 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.117006 (rank : 55) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.114053 (rank : 56) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.110323 (rank : 57) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.109281 (rank : 58) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.107204 (rank : 59) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
TPTE_HUMAN
|
||||||
| NC score | 0.095318 (rank : 60) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.093793 (rank : 61) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.092997 (rank : 62) | θ value | 0.163984 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.085543 (rank : 63) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.085156 (rank : 64) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.074708 (rank : 65) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.073353 (rank : 66) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.072293 (rank : 67) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.072270 (rank : 68) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.071452 (rank : 69) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.070580 (rank : 70) | θ value | 0.0148317 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.070510 (rank : 71) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.069219 (rank : 72) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.066469 (rank : 73) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.065848 (rank : 74) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.065355 (rank : 75) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.064453 (rank : 76) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.064433 (rank : 77) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.064273 (rank : 78) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.063593 (rank : 79) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.061166 (rank : 80) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.061050 (rank : 81) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.059692 (rank : 82) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.058675 (rank : 83) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.058212 (rank : 84) | θ value | 0.163984 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.057290 (rank : 85) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.056032 (rank : 86) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.051515 (rank : 87) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.051130 (rank : 88) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.050356 (rank : 89) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
SP17_MOUSE
|
||||||
| NC score | 0.045655 (rank : 90) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62252 | Gene names | Spa17, Sp17 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.042478 (rank : 91) | θ value | 0.62314 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
PSN2_MOUSE
|
||||||
| NC score | 0.033352 (rank : 92) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61144, O54977, P97934, P97935, Q91VS3, Q9D616 | Gene names | Psen2, Ad4h, Alg3, Ps-2, Psnl2 | |||
|
Domain Architecture |

|
|||||
| Description | Presenilin-2 (EC 3.4.23.-) (PS-2) [Contains: Presenilin-2 NTF subunit; Presenilin-2 CTF subunit]. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.031866 (rank : 93) | θ value | 0.0113563 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.028676 (rank : 94) | θ value | 0.0252991 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.027969 (rank : 95) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
IMMT_HUMAN
|
||||||
| NC score | 0.025932 (rank : 96) | θ value | 0.0563607 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16891, Q14539, Q15092, Q68D41, Q69HW5, Q6IBL0, Q7Z3X1, Q8TAJ5, Q9P0V2 | Gene names | IMMT, HMP, PIG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin) (p87/89) (Proliferation-inducing gene 4 protein). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.023028 (rank : 97) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.020936 (rank : 98) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.019847 (rank : 99) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.019823 (rank : 100) | θ value | 0.0736092 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.019650 (rank : 101) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.019225 (rank : 102) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
LC7L2_MOUSE
|
||||||
| NC score | 0.018661 (rank : 103) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.018526 (rank : 104) | θ value | 0.0252991 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
LC7L2_HUMAN
|
||||||
| NC score | 0.018063 (rank : 105) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
GLE1_MOUSE
|
||||||
| NC score | 0.017782 (rank : 106) | θ value | 0.47712 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.017137 (rank : 107) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
TEX9_MOUSE
|
||||||
| NC score | 0.015784 (rank : 108) | θ value | 0.813845 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.013542 (rank : 109) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
NELF_MOUSE
|
||||||
| NC score | 0.013172 (rank : 110) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99NF2, Q8BPT0, Q8C9R5, Q9DBF4, Q9ERZ1 | Gene names | Nelf, Jac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor) (Juxtasynaptic attractor of caldendrin on dendritic boutons protein) (Jacob protein). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.012962 (rank : 111) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.012812 (rank : 112) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
FA61A_HUMAN
|
||||||
| NC score | 0.012788 (rank : 113) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8ND56, Q76LX7, Q96AR3, Q96K73, Q96SN5, Q9UFR3 | Gene names | FAM61A, C19orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A (Putative alpha synuclein-binding protein) (AlphaSNBP). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.012687 (rank : 114) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.012497 (rank : 115) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.012291 (rank : 116) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.012113 (rank : 117) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.011959 (rank : 118) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.011844 (rank : 119) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
A16L1_MOUSE
|
||||||
| NC score | 0.011626 (rank : 120) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C0J2, Q6KAT7, Q80U97, Q80U98, Q80U99, Q80Y53, Q9DB63 | Gene names | Atg16l1, Apg16l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
NELF_HUMAN
|
||||||
| NC score | 0.011547 (rank : 121) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6X4W1, Q6X4V7, Q6X4V8, Q6X4V9, Q8N2M2, Q96SY1, Q9NPM4, Q9NPP3, Q9NPS3 | Gene names | NELF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.011151 (rank : 122) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.010528 (rank : 123) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
MCA1_HUMAN
|
||||||
| NC score | 0.010422 (rank : 124) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12904, Q6FG28, Q96CQ9 | Gene names | SCYE1, EMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.010172 (rank : 125) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.010018 (rank : 126) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.010017 (rank : 127) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.010016 (rank : 128) | θ value | 0.0736092 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.009938 (rank : 129) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.008748 (rank : 130) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.008594 (rank : 131) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
AR13B_HUMAN
|
||||||
| NC score | 0.008457 (rank : 132) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
MCA1_MOUSE
|
||||||
| NC score | 0.008452 (rank : 133) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P31230, Q60659 | Gene names | Scye1, Emap2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.008440 (rank : 134) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GP73_MOUSE
|
||||||
| NC score | 0.008392 (rank : 135) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
BFAR_HUMAN
|
||||||
| NC score | 0.008257 (rank : 136) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZS9 | Gene names | BFAR, BAR, RNF47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator (RING finger protein 47). | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.008232 (rank : 137) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
BFAR_MOUSE
|
||||||
| NC score | 0.008231 (rank : 138) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.007857 (rank : 139) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.007564 (rank : 140) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.007260 (rank : 141) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
TMCC3_HUMAN
|
||||||
| NC score | 0.006725 (rank : 142) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULS5, Q8IWB2 | Gene names | TMCC3, KIAA1145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 3. | |||||
|
ZWINT_HUMAN
|
||||||
| NC score | 0.006577 (rank : 143) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95229, Q9BWD0 | Gene names | ZWINT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
A16L1_HUMAN
|
||||||
| NC score | 0.006517 (rank : 144) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q676U5, Q6IPN1, Q6UXW4, Q6ZVZ5, Q8NCY2, Q96JV5, Q9H619 | Gene names | ATG16L1, APG16L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.006381 (rank : 145) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
NELFE_MOUSE
|
||||||
| NC score | 0.006308 (rank : 146) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19426, P97485, P97486, Q9CZN3 | Gene names | Rdbp, D17h6s45, Nelfe, Rd | |||
|
Domain Architecture |

|
|||||
| Description | Negative elongation factor E (NELF-E) (RD protein). | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.006306 (rank : 147) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
LAMC2_HUMAN
|
||||||
| NC score | 0.006295 (rank : 148) | θ value | 0.21417 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.006288 (rank : 149) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.006196 (rank : 150) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.005875 (rank : 151) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.005858 (rank : 152) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.005189 (rank : 153) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.005076 (rank : 154) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.004986 (rank : 155) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
ARD1H_HUMAN
|
||||||
| NC score | 0.004971 (rank : 156) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41227 | Gene names | ARD1A, ARD1, TE2 | |||
|
Domain Architecture |

|
|||||
| Description | N-terminal acetyltransferase complex ARD1 subunit homolog A (EC 2.3.1.88) (EC 2.3.1.-). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.004327 (rank : 157) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.004257 (rank : 158) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.003911 (rank : 159) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
NPC1_HUMAN
|
||||||
| NC score | 0.002736 (rank : 160) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15118, Q9P130 | Gene names | NPC1 | |||
|
Domain Architecture |

|
|||||
| Description | Niemann-Pick C1 protein precursor. | |||||
|
PML_HUMAN
|
||||||
| NC score | 0.002362 (rank : 161) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29590, P29591, P29592, P29593, Q00755 | Gene names | PML, MYL, RNF71, TRIM19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.002135 (rank : 162) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.001822 (rank : 163) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
OLF9_MOUSE
|
||||||
| NC score | -0.001729 (rank : 164) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60885, Q8VGC0 | Gene names | Olfr9, Mor269-3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 9 (Olfactory receptor 269-3) (Odorant receptor M25). | |||||
|
LHX3_HUMAN
|
||||||
| NC score | -0.002085 (rank : 165) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
MARK2_MOUSE
|
||||||
| NC score | -0.002525 (rank : 166) | θ value | 0.163984 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | -0.003537 (rank : 167) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
GALR1_HUMAN
|
||||||
| NC score | -0.003647 (rank : 168) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47211 | Gene names | GALR1, GALNR, GALNR1 | |||
|
Domain Architecture |

|
|||||
| Description | Galanin receptor type 1 (GAL1-R) (GALR1). | |||||
|
MARK2_HUMAN
|
||||||
| NC score | -0.003793 (rank : 169) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||