Please be patient as the page loads
|
KCNB1_HUMAN
|
||||||
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCNB1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 169 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998316 (rank : 2) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.992590 (rank : 3) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 5.11853e-104 (rank : 4) | NC score | 0.979864 (rank : 5) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 1.14028e-103 (rank : 5) | NC score | 0.979734 (rank : 6) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 5.29683e-101 (rank : 6) | NC score | 0.979291 (rank : 7) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 3.32542e-95 (rank : 7) | NC score | 0.979145 (rank : 8) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 3.32542e-95 (rank : 8) | NC score | 0.977882 (rank : 9) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 6.48998e-91 (rank : 9) | NC score | 0.980936 (rank : 4) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | 4.20666e-90 (rank : 10) | NC score | 0.971191 (rank : 11) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 3.33315e-87 (rank : 11) | NC score | 0.974746 (rank : 10) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | 2.82168e-86 (rank : 12) | NC score | 0.967183 (rank : 14) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | 3.94648e-80 (rank : 13) | NC score | 0.965025 (rank : 15) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | 5.8972e-76 (rank : 14) | NC score | 0.968083 (rank : 12) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | 1.71583e-75 (rank : 15) | NC score | 0.967615 (rank : 13) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 1.7756e-72 (rank : 16) | NC score | 0.955135 (rank : 16) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 2.31901e-72 (rank : 17) | NC score | 0.955018 (rank : 17) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 3.95564e-72 (rank : 18) | NC score | 0.950836 (rank : 18) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 7.45998e-71 (rank : 19) | NC score | 0.950129 (rank : 19) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | 2.24614e-67 (rank : 20) | NC score | 0.939465 (rank : 26) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | 2.93355e-67 (rank : 21) | NC score | 0.939110 (rank : 27) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | 8.53533e-67 (rank : 22) | NC score | 0.936358 (rank : 28) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | 2.4834e-66 (rank : 23) | NC score | 0.936052 (rank : 30) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 9.43692e-66 (rank : 24) | NC score | 0.942921 (rank : 20) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 1.2325e-65 (rank : 25) | NC score | 0.942920 (rank : 21) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 4.68348e-65 (rank : 26) | NC score | 0.941353 (rank : 22) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 6.11685e-65 (rank : 27) | NC score | 0.940741 (rank : 25) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | 1.50663e-63 (rank : 28) | NC score | 0.936278 (rank : 29) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 1.50663e-63 (rank : 29) | NC score | 0.940741 (rank : 24) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 1.96772e-63 (rank : 30) | NC score | 0.941197 (rank : 23) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | 2.56992e-63 (rank : 31) | NC score | 0.934734 (rank : 35) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | 5.7252e-63 (rank : 32) | NC score | 0.935696 (rank : 32) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | 7.47731e-63 (rank : 33) | NC score | 0.934880 (rank : 34) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | 1.11734e-58 (rank : 34) | NC score | 0.935582 (rank : 33) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | 1.90589e-58 (rank : 35) | NC score | 0.932879 (rank : 38) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | 2.48917e-58 (rank : 36) | NC score | 0.935797 (rank : 31) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | 2.7521e-57 (rank : 37) | NC score | 0.933294 (rank : 37) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 1.72781e-51 (rank : 38) | NC score | 0.934117 (rank : 36) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 6.56568e-51 (rank : 39) | NC score | 0.931808 (rank : 39) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 40) | NC score | 0.721307 (rank : 42) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 41) | NC score | 0.712009 (rank : 44) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 1.38178e-16 (rank : 42) | NC score | 0.719018 (rank : 43) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 43) | NC score | 0.710816 (rank : 45) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 44) | NC score | 0.749933 (rank : 40) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 45) | NC score | 0.743381 (rank : 41) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 46) | NC score | 0.680604 (rank : 47) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 47) | NC score | 0.681425 (rank : 46) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 48) | NC score | 0.489406 (rank : 49) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 49) | NC score | 0.490308 (rank : 48) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 50) | NC score | 0.244408 (rank : 71) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 51) | NC score | 0.204607 (rank : 86) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 52) | NC score | 0.242121 (rank : 72) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 53) | NC score | 0.213791 (rank : 81) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 54) | NC score | 0.246281 (rank : 70) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 55) | NC score | 0.247661 (rank : 69) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
KCTD2_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 56) | NC score | 0.297942 (rank : 54) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14681 | Gene names | KCTD2, KIAA0176 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 57) | NC score | 0.188129 (rank : 106) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 58) | NC score | 0.197172 (rank : 95) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 59) | NC score | 0.196508 (rank : 97) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 60) | NC score | 0.201413 (rank : 92) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 61) | NC score | 0.189575 (rank : 103) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
KCNN4_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 62) | NC score | 0.346998 (rank : 51) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCNN4_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 63) | NC score | 0.348681 (rank : 50) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCTD1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 64) | NC score | 0.304030 (rank : 52) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q719H9 | Gene names | KCTD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 65) | NC score | 0.304030 (rank : 53) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5M956, Q5EER9 | Gene names | Kctd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 66) | NC score | 0.211826 (rank : 83) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 67) | NC score | 0.194333 (rank : 99) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
KCTD5_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 68) | NC score | 0.274989 (rank : 59) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NXV2 | Gene names | KCTD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCTD5_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 69) | NC score | 0.276269 (rank : 58) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VC57 | Gene names | Kctd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 70) | NC score | 0.193642 (rank : 101) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 71) | NC score | 0.193723 (rank : 100) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 72) | NC score | 0.196531 (rank : 96) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 73) | NC score | 0.202262 (rank : 90) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 74) | NC score | 0.207453 (rank : 85) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 75) | NC score | 0.208153 (rank : 84) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
KCD17_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 76) | NC score | 0.224514 (rank : 79) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N5Z5, O95517 | Gene names | KCTD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD17. | |||||
|
KCNN1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 77) | NC score | 0.271492 (rank : 64) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 78) | NC score | 0.272626 (rank : 62) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 79) | NC score | 0.228577 (rank : 76) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 80) | NC score | 0.195660 (rank : 98) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 81) | NC score | 0.202522 (rank : 89) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
KCNN2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 82) | NC score | 0.280400 (rank : 56) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 83) | NC score | 0.280395 (rank : 57) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 84) | NC score | 0.273534 (rank : 61) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 85) | NC score | 0.189933 (rank : 102) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 86) | NC score | 0.202215 (rank : 91) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 87) | NC score | 0.274804 (rank : 60) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 88) | NC score | 0.229663 (rank : 74) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 89) | NC score | 0.203883 (rank : 88) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
KCD15_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 90) | NC score | 0.284933 (rank : 55) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96SI1, Q9BVI6 | Gene names | KCTD15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
WDR67_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 91) | NC score | 0.010441 (rank : 151) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0.125558 (rank : 92) | NC score | 0.197253 (rank : 94) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 93) | NC score | 0.010084 (rank : 153) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
KCNT1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 94) | NC score | 0.224882 (rank : 78) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 95) | NC score | 0.014007 (rank : 146) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
CING_MOUSE
|
||||||
| θ value | 0.279714 (rank : 96) | NC score | 0.000573 (rank : 168) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
KCD14_HUMAN
|
||||||
| θ value | 0.279714 (rank : 97) | NC score | 0.184633 (rank : 107) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BQ13 | Gene names | KCTD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD14. | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 98) | NC score | 0.204027 (rank : 87) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNRG_HUMAN
|
||||||
| θ value | 0.279714 (rank : 99) | NC score | 0.229185 (rank : 75) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8N5I3, Q8IU75, Q8N3Q9 | Gene names | KCNRG, CLLD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 0.365318 (rank : 100) | NC score | 0.188591 (rank : 105) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 0.365318 (rank : 101) | NC score | 0.189109 (rank : 104) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | 0.365318 (rank : 102) | NC score | 0.178042 (rank : 108) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCTD2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 103) | NC score | 0.151404 (rank : 114) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CEZ0 | Gene names | Kctd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
KCD15_MOUSE
|
||||||
| θ value | 0.47712 (rank : 104) | NC score | 0.272617 (rank : 63) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K0E1 | Gene names | Kctd15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCTD3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 105) | NC score | 0.240069 (rank : 73) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y597, Q49AG7, Q504Q9, Q6PJN6, Q8ND58, Q8NDJ0, Q8WX16 | Gene names | KCTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3 (NY-REN-45 antigen). | |||||
|
KCTD3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 106) | NC score | 0.226934 (rank : 77) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BFX3, Q6P7X4 | Gene names | Kctd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3. | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 107) | NC score | 0.013943 (rank : 148) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 108) | NC score | 0.014003 (rank : 147) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
CLM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 109) | NC score | 0.015828 (rank : 144) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6SJQ7, Q6PEU7, Q8K4V9 | Gene names | Cd300lf, Clm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CMRF35-like-molecule 1 precursor (CLM-1) (CD300 antigen like family member F). | |||||
|
SNX1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 110) | NC score | 0.012559 (rank : 149) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV80, Q9EQZ9 | Gene names | Snx1 | |||
|
Domain Architecture |
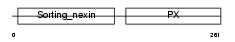
|
|||||
| Description | Sorting nexin-1. | |||||
|
KCNK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 111) | NC score | 0.053132 (rank : 137) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 112) | NC score | 0.130986 (rank : 120) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 113) | NC score | 0.134112 (rank : 119) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCD11_MOUSE
|
||||||
| θ value | 1.38821 (rank : 114) | NC score | 0.117149 (rank : 122) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K485 | Gene names | Kctd11, Ren | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 115) | NC score | 0.030353 (rank : 143) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCTD4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 116) | NC score | 0.249638 (rank : 68) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WVF5 | Gene names | KCTD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 117) | NC score | 0.250663 (rank : 67) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9D7X1, Q8CCQ3, Q9CYK4 | Gene names | Kctd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 118) | NC score | 0.002224 (rank : 164) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ORC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 119) | NC score | 0.014407 (rank : 145) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13415, Q13471, Q5T0F5 | Gene names | ORC1L, ORC1, PARC1 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 1 (Replication control protein 1). | |||||
|
PO4F1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 120) | NC score | 0.003667 (rank : 161) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17208 | Gene names | Pou4f1, Brn-3, Brn3, Brn3a | |||
|
Domain Architecture |
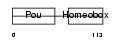
|
|||||
| Description | POU domain, class 4, transcription factor 1 (Brain-specific homeobox/POU domain protein 3A) (Brn-3A) (Brn-3.0). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 121) | NC score | 0.010252 (rank : 152) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
HLX1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 122) | NC score | 0.000182 (rank : 170) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61670 | Gene names | Hlx1, Hlx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein HLX1. | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 123) | NC score | 0.167848 (rank : 112) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 124) | NC score | 0.009527 (rank : 154) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
KCTD9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 125) | NC score | 0.169979 (rank : 111) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7L273, Q6NUM8, Q9NXV4 | Gene names | KCTD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
KCTD9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 126) | NC score | 0.174055 (rank : 110) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80UN1 | Gene names | Kctd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 127) | NC score | -0.001197 (rank : 175) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 128) | NC score | 0.003978 (rank : 160) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 3.0926 (rank : 129) | NC score | -0.004433 (rank : 180) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 130) | NC score | 0.041963 (rank : 142) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 131) | NC score | 0.041985 (rank : 141) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 132) | NC score | 0.152028 (rank : 113) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNRG_MOUSE
|
||||||
| θ value | 3.0926 (rank : 133) | NC score | 0.212310 (rank : 82) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q2TUM3, Q6P8J6, Q8C6V1 | Gene names | Kcnrg, Clld4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
KCTD6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 134) | NC score | 0.260637 (rank : 66) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8NC69, Q8NBS6, Q8TCA6 | Gene names | KCTD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 135) | NC score | 0.262908 (rank : 65) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BNL5, Q3TXX1 | Gene names | Kctd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 136) | NC score | -0.000679 (rank : 172) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PO4F1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 137) | NC score | 0.003232 (rank : 162) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01851, Q14986, Q15318 | Gene names | POU4F1, BRN3A, RDC1 | |||
|
Domain Architecture |
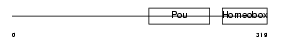
|
|||||
| Description | POU domain, class 4, transcription factor 1 (Brain-specific homeobox/POU domain protein 3A) (Brn-3A) (Oct-T1) (Homeobox/POU domain protein RDC-1). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 138) | NC score | 0.042453 (rank : 139) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 139) | NC score | 0.042421 (rank : 140) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 140) | NC score | 0.136416 (rank : 118) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6ZPR4, Q8C3E7 | Gene names | Kcnt1, Kiaa1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1. | |||||
|
MNDA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 141) | NC score | 0.011729 (rank : 150) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |
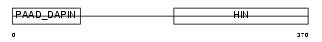
|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
PAK3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 142) | NC score | -0.007053 (rank : 181) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75914, Q5JWX1, Q5JWX2, Q7Z2D6, Q7Z2E4, Q7Z3Z8, Q8WWK5, Q8WX23, Q9P0J8 | Gene names | PAK3, OPHN3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 3 (EC 2.7.11.1) (p21-activated kinase 3) (PAK-3) (Beta-PAK) (Oligophrenin-3). | |||||
|
PHF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 143) | NC score | 0.002914 (rank : 163) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 4.03905 (rank : 144) | NC score | -0.000960 (rank : 173) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
HXA6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | -0.001837 (rank : 176) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09092, Q9JI98 | Gene names | Hoxa6, Hox-1.2, Hoxa-6 | |||
|
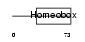
Domain Architecture |
|
|||||
| Description | Homeobox protein Hox-A6 (Hox-1.2) (M5-4). | |||||
|
KCNK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.051080 (rank : 138) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 147) | NC score | 0.139483 (rank : 116) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.000450 (rank : 169) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CU025_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.006571 (rank : 155) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y426, Q5R2V7, Q6AHX8, Q9NSE6 | Gene names | C21orf25, C21orf258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf25 precursor. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | -0.004004 (rank : 179) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KCD11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.110391 (rank : 123) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q693B1 | Gene names | KCTD11, REN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.141716 (rank : 115) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.086176 (rank : 129) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCTD7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.223402 (rank : 80) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BJK1, Q3USA6, Q8C0S7 | Gene names | Kctd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | -0.001178 (rank : 174) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
MMRN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | -0.002162 (rank : 177) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13201, Q6P3T8 | Gene names | MMRN1, ECM, EMILIN4, MMRN | |||
|
Domain Architecture |

|
|||||
| Description | Multimerin-1 precursor (Endothelial cell multimerin 1) (EMILIN-4) (Elastin microfibril interface located protein 4) (Elastin microfibril interfacer 4). | |||||
|
PDE3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.001373 (rank : 166) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
SETD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.005717 (rank : 156) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.005692 (rank : 157) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
UNC5A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | -0.000175 (rank : 171) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1S4, Q6PEF7, Q80T71 | Gene names | Unc5a, Kiaa1976, Unc5h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5A precursor (Unc-5 homolog A) (Unc-5 homolog 1). | |||||
|
ZF106_HUMAN
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.000909 (rank : 167) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
ABCC8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.001868 (rank : 165) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q09428, O75948, Q16583 | Gene names | ABCC8, SUR, SUR1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette transporter sub-family C member 8 (Sulfonylurea receptor 1). | |||||
|
DEMA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.004471 (rank : 159) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08495, Q13215, Q9BRE3 | Gene names | EPB49, DMT | |||
|
Domain Architecture |

|
|||||
| Description | Dematin (Erythrocyte membrane protein band 4.9). | |||||
|
DEMA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.004538 (rank : 158) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV69 | Gene names | Epb49, Epb4.9 | |||
|
Domain Architecture |

|
|||||
| Description | Dematin (Erythrocyte membrane protein band 4.9). | |||||
|
KCNK9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.104781 (rank : 124) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.102697 (rank : 125) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | -0.003267 (rank : 178) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PAK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | -0.007344 (rank : 182) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61036, O88645, Q8K1R5, Q8K1R6 | Gene names | Pak3, Pak-3, Pakb, Stk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 3 (EC 2.7.11.1) (p21-activated kinase 3) (PAK-3) (Beta-PAK) (CDC42/RAC effector kinase PAK-B). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.176887 (rank : 109) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
KCD12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.081581 (rank : 131) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96CX2 | Gene names | KCTD12, C13orf2, KIAA1778, PFET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
KCD12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.076860 (rank : 132) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6WVG3 | Gene names | Kctd12, Pfet1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
KCD16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.082951 (rank : 130) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q68DU8, Q9P2M9 | Gene names | KCTD16, KIAA1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCD16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.087218 (rank : 128) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5DTY9, Q78Y50 | Gene names | Kctd16, Kiaa1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCD18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.053631 (rank : 136) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.138560 (rank : 117) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.065028 (rank : 135) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.123230 (rank : 121) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.069828 (rank : 134) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.073697 (rank : 133) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCTD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.198759 (rank : 93) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96MP8, Q8IVR0 | Gene names | KCTD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
KCTD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.092430 (rank : 127) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6ZWB6 | Gene names | KCTD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCTD8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.102234 (rank : 126) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q50H33, Q8BR74, Q8C4C2, Q8C906, Q8C9B0, Q8CAA9 | Gene names | Kctd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 169 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.998316 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.992590 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.980936 (rank : 4) | θ value | 6.48998e-91 (rank : 9) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.979864 (rank : 5) | θ value | 5.11853e-104 (rank : 4) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.979734 (rank : 6) | θ value | 1.14028e-103 (rank : 5) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.979291 (rank : 7) | θ value | 5.29683e-101 (rank : 6) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.979145 (rank : 8) | θ value | 3.32542e-95 (rank : 7) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.977882 (rank : 9) | θ value | 3.32542e-95 (rank : 8) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.974746 (rank : 10) | θ value | 3.33315e-87 (rank : 11) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.971191 (rank : 11) | θ value | 4.20666e-90 (rank : 10) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.968083 (rank : 12) | θ value | 5.8972e-76 (rank : 14) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.967615 (rank : 13) | θ value | 1.71583e-75 (rank : 15) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.967183 (rank : 14) | θ value | 2.82168e-86 (rank : 12) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.965025 (rank : 15) | θ value | 3.94648e-80 (rank : 13) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.955135 (rank : 16) | θ value | 1.7756e-72 (rank : 16) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.955018 (rank : 17) | θ value | 2.31901e-72 (rank : 17) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.950836 (rank : 18) | θ value | 3.95564e-72 (rank : 18) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.950129 (rank : 19) | θ value | 7.45998e-71 (rank : 19) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.942921 (rank : 20) | θ value | 9.43692e-66 (rank : 24) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.942920 (rank : 21) | θ value | 1.2325e-65 (rank : 25) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.941353 (rank : 22) | θ value | 4.68348e-65 (rank : 26) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.941197 (rank : 23) | θ value | 1.96772e-63 (rank : 30) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.940741 (rank : 24) | θ value | 1.50663e-63 (rank : 29) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.940741 (rank : 25) | θ value | 6.11685e-65 (rank : 27) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.939465 (rank : 26) | θ value | 2.24614e-67 (rank : 20) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.939110 (rank : 27) | θ value | 2.93355e-67 (rank : 21) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.936358 (rank : 28) | θ value | 8.53533e-67 (rank : 22) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.936278 (rank : 29) | θ value | 1.50663e-63 (rank : 28) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.936052 (rank : 30) | θ value | 2.4834e-66 (rank : 23) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.935797 (rank : 31) | θ value | 2.48917e-58 (rank : 36) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.935696 (rank : 32) | θ value | 5.7252e-63 (rank : 32) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.935582 (rank : 33) | θ value | 1.11734e-58 (rank : 34) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.934880 (rank : 34) | θ value | 7.47731e-63 (rank : 33) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.934734 (rank : 35) | θ value | 2.56992e-63 (rank : 31) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.934117 (rank : 36) | θ value | 1.72781e-51 (rank : 38) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.933294 (rank : 37) | θ value | 2.7521e-57 (rank : 37) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.932879 (rank : 38) | θ value | 1.90589e-58 (rank : 35) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.931808 (rank : 39) | θ value | 6.56568e-51 (rank : 39) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.749933 (rank : 40) | θ value | 2.35696e-16 (rank : 44) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.743381 (rank : 41) | θ value | 8.95645e-16 (rank : 45) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.721307 (rank : 42) | θ value | 2.7842e-17 (rank : 40) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.719018 (rank : 43) | θ value | 1.38178e-16 (rank : 42) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.712009 (rank : 44) | θ value | 4.74913e-17 (rank : 41) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.710816 (rank : 45) | θ value | 1.38178e-16 (rank : 43) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.681425 (rank : 46) | θ value | 1.68911e-14 (rank : 47) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.680604 (rank : 47) | θ value | 1.68911e-14 (rank : 46) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.490308 (rank : 48) | θ value | 6.21693e-09 (rank : 49) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.489406 (rank : 49) | θ value | 6.21693e-09 (rank : 48) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCNN4_MOUSE
|
||||||
| NC score | 0.348681 (rank : 50) | θ value | 0.000461057 (rank : 63) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNN4_HUMAN
|
||||||
| NC score | 0.346998 (rank : 51) | θ value | 0.000461057 (rank : 62) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCTD1_HUMAN
|
||||||
| NC score | 0.304030 (rank : 52) | θ value | 0.000786445 (rank : 64) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q719H9 | Gene names | KCTD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD1_MOUSE
|
||||||
| NC score | 0.304030 (rank : 53) | θ value | 0.000786445 (rank : 65) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5M956, Q5EER9 | Gene names | Kctd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD2_HUMAN
|
||||||
| NC score | 0.297942 (rank : 54) | θ value | 6.43352e-06 (rank : 56) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14681 | Gene names | KCTD2, KIAA0176 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
KCD15_HUMAN
|
||||||
| NC score | 0.284933 (rank : 55) | θ value | 0.0736092 (rank : 90) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96SI1, Q9BVI6 | Gene names | KCTD15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCNN2_HUMAN
|
||||||
| NC score | 0.280400 (rank : 56) | θ value | 0.0252991 (rank : 82) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.280395 (rank : 57) | θ value | 0.0252991 (rank : 83) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCTD5_MOUSE
|
||||||
| NC score | 0.276269 (rank : 58) | θ value | 0.00175202 (rank : 69) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VC57 | Gene names | Kctd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCTD5_HUMAN
|
||||||
| NC score | 0.274989 (rank : 59) | θ value | 0.00134147 (rank : 68) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NXV2 | Gene names | KCTD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.274804 (rank : 60) | θ value | 0.0330416 (rank : 87) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.273534 (rank : 61) | θ value | 0.0252991 (rank : 84) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
KCNN1_MOUSE
|
||||||
| NC score | 0.272626 (rank : 62) | θ value | 0.0193708 (rank : 78) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCD15_MOUSE
|
||||||
| NC score | 0.272617 (rank : 63) | θ value | 0.47712 (rank : 104) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K0E1 | Gene names | Kctd15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCNN1_HUMAN
|
||||||
| NC score | 0.271492 (rank : 64) | θ value | 0.0193708 (rank : 77) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCTD6_MOUSE
|
||||||
| NC score | 0.262908 (rank : 65) | θ value | 3.0926 (rank : 135) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BNL5, Q3TXX1 | Gene names | Kctd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD6_HUMAN
|
||||||
| NC score | 0.260637 (rank : 66) | θ value | 3.0926 (rank : 134) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8NC69, Q8NBS6, Q8TCA6 | Gene names | KCTD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD4_MOUSE
|
||||||
| NC score | 0.250663 (rank : 67) | θ value | 1.38821 (rank : 117) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9D7X1, Q8CCQ3, Q9CYK4 | Gene names | Kctd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD4_HUMAN
|
||||||
| NC score | 0.249638 (rank : 68) | θ value | 1.38821 (rank : 116) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WVF5 | Gene names | KCTD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.247661 (rank : 69) | θ value | 3.77169e-06 (rank : 55) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.246281 (rank : 70) | θ value | 7.59969e-07 (rank : 54) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.244408 (rank : 71) | θ value | 3.41135e-07 (rank : 50) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.242121 (rank : 72) | θ value | 5.81887e-07 (rank : 52) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
KCTD3_HUMAN
|
||||||
| NC score | 0.240069 (rank : 73) | θ value | 0.47712 (rank : 105) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y597, Q49AG7, Q504Q9, Q6PJN6, Q8ND58, Q8NDJ0, Q8WX16 | Gene names | KCTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3 (NY-REN-45 antigen). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.229663 (rank : 74) | θ value | 0.0330416 (rank : 88) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
KCNRG_HUMAN
|
||||||
| NC score | 0.229185 (rank : 75) | θ value | 0.279714 (rank : 99) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8N5I3, Q8IU75, Q8N3Q9 | Gene names | KCNRG, CLLD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.228577 (rank : 76) | θ value | 0.0193708 (rank : 79) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
KCTD3_MOUSE
|
||||||
| NC score | 0.226934 (rank : 77) | θ value | 0.47712 (rank : 106) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BFX3, Q6P7X4 | Gene names | Kctd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3. | |||||
|
KCNT1_HUMAN
|
||||||
| NC score | 0.224882 (rank : 78) | θ value | 0.163984 (rank : 94) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KCD17_HUMAN
|
||||||
| NC score | 0.224514 (rank : 79) | θ value | 0.0148317 (rank : 76) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N5Z5, O95517 | Gene names | KCTD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD17. | |||||
|
KCTD7_MOUSE
|
||||||
| NC score | 0.223402 (rank : 80) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BJK1, Q3USA6, Q8C0S7 | Gene names | Kctd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.213791 (rank : 81) | θ value | 7.59969e-07 (rank : 53) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
KCNRG_MOUSE
|
||||||
| NC score | 0.212310 (rank : 82) | θ value | 3.0926 (rank : 133) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q2TUM3, Q6P8J6, Q8C6V1 | Gene names | Kcnrg, Clld4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.211826 (rank : 83) | θ value | 0.00102713 (rank : 66) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.208153 (rank : 84) | θ value | 0.0148317 (rank : 75) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.207453 (rank : 85) | θ value | 0.0148317 (rank : 74) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.204607 (rank : 86) | θ value | 5.81887e-07 (rank : 51) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.204027 (rank : 87) | θ value | 0.279714 (rank : 98) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.203883 (rank : 88) | θ value | 0.0431538 (rank : 89) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.202522 (rank : 89) | θ value | 0.0252991 (rank : 81) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.202262 (rank : 90) | θ value | 0.0113563 (rank : 73) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.202215 (rank : 91) | θ value | 0.0330416 (rank : 86) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.201413 (rank : 92) | θ value | 0.000270298 (rank : 60) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
KCTD7_HUMAN
|
||||||
| NC score | 0.198759 (rank : 93) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96MP8, Q8IVR0 | Gene names | KCTD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.197253 (rank : 94) | θ value | 0.125558 (rank : 92) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.197172 (rank : 95) | θ value | 4.1701e-05 (rank : 58) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.196531 (rank : 96) | θ value | 0.0113563 (rank : 72) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.196508 (rank : 97) | θ value | 0.000158464 (rank : 59) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.195660 (rank : 98) | θ value | 0.0193708 (rank : 80) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.194333 (rank : 99) | θ value | 0.00134147 (rank : 67) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.193723 (rank : 100) | θ value | 0.00228821 (rank : 71) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.193642 (rank : 101) | θ value | 0.00228821 (rank : 70) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.189933 (rank : 102) | θ value | 0.0252991 (rank : 85) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.189575 (rank : 103) | θ value | 0.00035302 (rank : 61) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.189109 (rank : 104) | θ value | 0.365318 (rank : 101) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.188591 (rank : 105) | θ value | 0.365318 (rank : 100) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.188129 (rank : 106) | θ value | 2.44474e-05 (rank : 57) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
KCD14_HUMAN
|
||||||
| NC score | 0.184633 (rank : 107) | θ value | 0.279714 (rank : 97) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BQ13 | Gene names | KCTD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD14. | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.178042 (rank : 108) | θ value | 0.365318 (rank : 102) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.176887 (rank : 109) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
KCTD9_MOUSE
|
||||||
| NC score | 0.174055 (rank : 110) | θ value | 2.36792 (rank : 126) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80UN1 | Gene names | Kctd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
KCTD9_HUMAN
|
||||||
| NC score | 0.169979 (rank : 111) | θ value | 2.36792 (rank : 125) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7L273, Q6NUM8, Q9NXV4 | Gene names | KCTD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.167848 (rank : 112) | θ value | 1.81305 (rank : 123) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.152028 (rank : 113) | θ value | 3.0926 (rank : 132) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCTD2_MOUSE
|
||||||
| NC score | 0.151404 (rank : 114) | θ value | 0.365318 (rank : 103) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CEZ0 | Gene names | Kctd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.141716 (rank : 115) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.139483 (rank : 116) | θ value | 5.27518 (rank : 147) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.138560 (rank : 117) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNT1_MOUSE
|
||||||
| NC score | 0.136416 (rank : 118) | θ value | 4.03905 (rank : 140) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6ZPR4, Q8C3E7 | Gene names | Kcnt1, Kiaa1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1. | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.134112 (rank : 119) | θ value | 1.06291 (rank : 113) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.130986 (rank : 120) | θ value | 1.06291 (rank : 112) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.123230 (rank : 121) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCD11_MOUSE
|
||||||
| NC score | 0.117149 (rank : 122) | θ value | 1.38821 (rank : 114) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K485 | Gene names | Kctd11, Ren | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCD11_HUMAN
|
||||||
| NC score | 0.110391 (rank : 123) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q693B1 | Gene names | KCTD11, REN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCNK9_HUMAN
|
||||||
| NC score | 0.104781 (rank : 124) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.102697 (rank : 125) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCTD8_MOUSE
|
||||||
| NC score | 0.102234 (rank : 126) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q50H33, Q8BR74, Q8C4C2, Q8C906, Q8C9B0, Q8CAA9 | Gene names | Kctd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCTD8_HUMAN
|
||||||
| NC score | 0.092430 (rank : 127) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6ZWB6 | Gene names | KCTD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCD16_MOUSE
|
||||||
| NC score | 0.087218 (rank : 128) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5DTY9, Q78Y50 | Gene names | Kctd16, Kiaa1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.086176 (rank : 129) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCD16_HUMAN
|
||||||
| NC score | 0.082951 (rank : 130) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q68DU8, Q9P2M9 | Gene names | KCTD16, KIAA1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCD12_HUMAN
|
||||||
| NC score | 0.081581 (rank : 131) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96CX2 | Gene names | KCTD12, C13orf2, KIAA1778, PFET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
KCD12_MOUSE
|
||||||
| NC score | 0.076860 (rank : 132) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6WVG3 | Gene names | Kctd12, Pfet1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.073697 (rank : 133) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNK7_HUMAN
|
||||||
| NC score | 0.069828 (rank : 134) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.065028 (rank : 135) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCD18_HUMAN
|
||||||
| NC score | 0.053631 (rank : 136) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
KCNK1_MOUSE
|
||||||
| NC score | 0.053132 (rank : 137) | θ value | 1.06291 (rank : 111) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNK1_HUMAN
|
||||||
| NC score | 0.051080 (rank : 138) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.042453 (rank : 139) | θ value | 4.03905 (rank : 138) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.042421 (rank : 140) | θ value | 4.03905 (rank : 139) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.041985 (rank : 141) | θ value | 3.0926 (rank : 131) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.041963 (rank : 142) | θ value | 3.0926 (rank : 130) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.030353 (rank : 143) | θ value | 1.38821 (rank : 115) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
CLM1_MOUSE
|
||||||
| NC score | 0.015828 (rank : 144) | θ value | 0.813845 (rank : 109) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6SJQ7, Q6PEU7, Q8K4V9 | Gene names | Cd300lf, Clm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CMRF35-like-molecule 1 precursor (CLM-1) (CD300 antigen like family member F). | |||||
|
ORC1_HUMAN
|
||||||
| NC score | 0.014407 (rank : 145) | θ value | 1.38821 (rank : 119) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13415, Q13471, Q5T0F5 | Gene names | ORC1L, ORC1, PARC1 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 1 (Replication control protein 1). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.014007 (rank : 146) | θ value | 0.21417 (rank : 95) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
RCOR2_MOUSE
|
||||||
| NC score | 0.014003 (rank : 147) | θ value | 0.62314 (rank : 108) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.013943 (rank : 148) | θ value | 0.62314 (rank : 107) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
SNX1_MOUSE
|
||||||
| NC score | 0.012559 (rank : 149) | θ value | 0.813845 (rank : 110) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV80, Q9EQZ9 | Gene names | Snx1 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-1. | |||||
|
MNDA_HUMAN
|
||||||
| NC score | 0.011729 (rank : 150) | θ value | 4.03905 (rank : 141) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
WDR67_HUMAN
|
||||||
| NC score | 0.010441 (rank : 151) | θ value | 0.0961366 (rank : 91) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.010252 (rank : 152) | θ value | 1.81305 (rank : 121) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.010084 (rank : 153) | θ value | 0.125558 (rank : 93) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.009527 (rank : 154) | θ value | 1.81305 (rank : 124) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CU025_HUMAN
|
||||||
| NC score | 0.006571 (rank : 155) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y426, Q5R2V7, Q6AHX8, Q9NSE6 | Gene names | C21orf25, C21orf258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf25 precursor. | |||||
|
SETD8_HUMAN
|
||||||
| NC score | 0.005717 (rank : 156) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.005692 (rank : 157) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
DEMA_MOUSE
|
||||||
| NC score | 0.004538 (rank : 158) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV69 | Gene names | Epb49, Epb4.9 | |||
|
Domain Architecture |

|
|||||
| Description | Dematin (Erythrocyte membrane protein band 4.9). | |||||
|
DEMA_HUMAN
|
||||||
| NC score | 0.004471 (rank : 159) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08495, Q13215, Q9BRE3 | Gene names | EPB49, DMT | |||
|
Domain Architecture |

|
|||||
| Description | Dematin (Erythrocyte membrane protein band 4.9). | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.003978 (rank : 160) | θ value | 3.0926 (rank : 128) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
PO4F1_MOUSE
|
||||||
| NC score | 0.003667 (rank : 161) | θ value | 1.38821 (rank : 120) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17208 | Gene names | Pou4f1, Brn-3, Brn3, Brn3a | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 1 (Brain-specific homeobox/POU domain protein 3A) (Brn-3A) (Brn-3.0). | |||||
|
PO4F1_HUMAN
|
||||||
| NC score | 0.003232 (rank : 162) | θ value | 3.0926 (rank : 137) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01851, Q14986, Q15318 | Gene names | POU4F1, BRN3A, RDC1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 1 (Brain-specific homeobox/POU domain protein 3A) (Brn-3A) (Oct-T1) (Homeobox/POU domain protein RDC-1). | |||||
|
PHF2_HUMAN
|
||||||
| NC score | 0.002914 (rank : 163) | θ value | 4.03905 (rank : 143) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.002224 (rank : 164) | θ value | 1.38821 (rank : 118) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ABCC8_HUMAN
|
||||||
| NC score | 0.001868 (rank : 165) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q09428, O75948, Q16583 | Gene names | ABCC8, SUR, SUR1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette transporter sub-family C member 8 (Sulfonylurea receptor 1). | |||||
|
PDE3A_HUMAN
|
||||||
| NC score | 0.001373 (rank : 166) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
ZF106_HUMAN
|
||||||
| NC score | 0.000909 (rank : 167) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.000573 (rank : 168) | θ value | 0.279714 (rank : 96) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.000450 (rank : 169) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
HLX1_MOUSE
|
||||||
| NC score | 0.000182 (rank : 170) | θ value | 1.81305 (rank : 122) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61670 | Gene names | Hlx1, Hlx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein HLX1. | |||||
|
UNC5A_MOUSE
|
||||||
| NC score | -0.000175 (rank : 171) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1S4, Q6PEF7, Q80T71 | Gene names | Unc5a, Kiaa1976, Unc5h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5A precursor (Unc-5 homolog A) (Unc-5 homolog 1). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | -0.000679 (rank : 172) | θ value | 3.0926 (rank : 136) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
ZN292_HUMAN
|
||||||
| NC score | -0.000960 (rank : 173) | θ value | 4.03905 (rank : 144) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
LDB3_MOUSE
|
||||||
| NC score | -0.001178 (rank : 174) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | -0.001197 (rank : 175) | θ value | 2.36792 (rank : 127) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
HXA6_MOUSE
|
||||||
| NC score | -0.001837 (rank : 176) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09092, Q9JI98 | Gene names | Hoxa6, Hox-1.2, Hoxa-6 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A6 (Hox-1.2) (M5-4). | |||||
|
MMRN1_HUMAN
|
||||||
| NC score | -0.002162 (rank : 177) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13201, Q6P3T8 | Gene names | MMRN1, ECM, EMILIN4, MMRN | |||
|
Domain Architecture |

|
|||||
| Description | Multimerin-1 precursor (Endothelial cell multimerin 1) (EMILIN-4) (Elastin microfibril interface located protein 4) (Elastin microfibril interfacer 4). | |||||
|
NFH_HUMAN
|
||||||
| NC score | -0.003267 (rank : 178) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
K1802_HUMAN
|
||||||
| NC score | -0.004004 (rank : 179) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CE290_HUMAN
|
||||||
| NC score | -0.004433 (rank : 180) | θ value | 3.0926 (rank : 129) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
PAK3_HUMAN
|
||||||
| NC score | -0.007053 (rank : 181) | θ value | 4.03905 (rank : 142) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75914, Q5JWX1, Q5JWX2, Q7Z2D6, Q7Z2E4, Q7Z3Z8, Q8WWK5, Q8WX23, Q9P0J8 | Gene names | PAK3, OPHN3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 3 (EC 2.7.11.1) (p21-activated kinase 3) (PAK-3) (Beta-PAK) (Oligophrenin-3). | |||||
|
PAK3_MOUSE
|
||||||
| NC score | -0.007344 (rank : 182) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61036, O88645, Q8K1R5, Q8K1R6 | Gene names | Pak3, Pak-3, Pakb, Stk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 3 (EC 2.7.11.1) (p21-activated kinase 3) (PAK-3) (Beta-PAK) (CDC42/RAC effector kinase PAK-B). | |||||