Please be patient as the page loads
|
CAC1F_HUMAN
|
||||||
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAC1A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.947736 (rank : 13) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.955464 (rank : 11) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.956055 (rank : 10) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.951368 (rank : 12) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.991536 (rank : 7) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.991918 (rank : 6) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.993397 (rank : 5) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.960135 (rank : 9) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.960243 (rank : 8) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.997447 (rank : 2) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.993545 (rank : 3) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 0 (rank : 13) | NC score | 0.993419 (rank : 4) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 6.7473e-72 (rank : 14) | NC score | 0.858226 (rank : 18) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 4.83544e-70 (rank : 15) | NC score | 0.840279 (rank : 25) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 6.31527e-70 (rank : 16) | NC score | 0.896416 (rank : 14) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 1.83746e-69 (rank : 17) | NC score | 0.843151 (rank : 23) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 3.13423e-69 (rank : 18) | NC score | 0.840048 (rank : 26) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 4.52582e-68 (rank : 19) | NC score | 0.850337 (rank : 21) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 1.00825e-67 (rank : 20) | NC score | 0.859965 (rank : 17) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 6.53529e-67 (rank : 21) | NC score | 0.839791 (rank : 27) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 3.24342e-66 (rank : 22) | NC score | 0.837100 (rank : 29) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 3.58603e-65 (rank : 23) | NC score | 0.867686 (rank : 16) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 3.58603e-65 (rank : 24) | NC score | 0.870875 (rank : 15) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 1.04338e-64 (rank : 25) | NC score | 0.842081 (rank : 24) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 6.76295e-64 (rank : 26) | NC score | 0.852519 (rank : 20) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 1.15359e-63 (rank : 27) | NC score | 0.852978 (rank : 19) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 1.50663e-63 (rank : 28) | NC score | 0.837717 (rank : 28) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 7.47731e-63 (rank : 29) | NC score | 0.837087 (rank : 30) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 1.78386e-56 (rank : 30) | NC score | 0.849894 (rank : 22) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 31) | NC score | 0.255439 (rank : 36) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 32) | NC score | 0.392289 (rank : 31) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 33) | NC score | 0.339065 (rank : 35) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 34) | NC score | 0.355427 (rank : 33) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 35) | NC score | 0.358209 (rank : 32) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 36) | NC score | 0.344946 (rank : 34) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 37) | NC score | 0.240819 (rank : 37) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 38) | NC score | 0.191010 (rank : 44) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 39) | NC score | 0.191135 (rank : 43) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 40) | NC score | 0.187757 (rank : 45) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 41) | NC score | 0.210964 (rank : 40) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 42) | NC score | 0.214593 (rank : 39) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 43) | NC score | 0.184736 (rank : 47) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 44) | NC score | 0.184874 (rank : 46) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 45) | NC score | 0.207453 (rank : 41) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 46) | NC score | 0.207381 (rank : 42) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 47) | NC score | 0.220106 (rank : 38) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 48) | NC score | 0.150767 (rank : 51) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 49) | NC score | 0.141107 (rank : 54) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 50) | NC score | 0.130541 (rank : 56) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 51) | NC score | 0.161656 (rank : 50) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 52) | NC score | 0.018203 (rank : 103) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 53) | NC score | 0.128960 (rank : 58) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 54) | NC score | 0.169855 (rank : 49) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 55) | NC score | 0.028378 (rank : 92) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 56) | NC score | 0.117896 (rank : 60) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
ACM5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 57) | NC score | 0.004814 (rank : 138) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08912, Q96RG7 | Gene names | CHRM5 | |||
|
Domain Architecture |
|
|||||
| Description | Muscarinic acetylcholine receptor M5. | |||||
|
BSPRY_MOUSE
|
||||||
| θ value | 0.125558 (rank : 58) | NC score | 0.018101 (rank : 104) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YW5, Q3TU74, Q8BZF0, Q99KV7, Q9ER70 | Gene names | Bspry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B box and SPRY domain-containing protein. | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 59) | NC score | 0.173619 (rank : 48) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
ACM1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 60) | NC score | 0.004509 (rank : 140) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 917 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P12657, Q8BJN3 | Gene names | Chrm1, Chrm-1 | |||
|
Domain Architecture |
|
|||||
| Description | Muscarinic acetylcholine receptor M1. | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 61) | NC score | 0.132833 (rank : 55) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 62) | NC score | 0.119194 (rank : 59) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 63) | NC score | 0.013648 (rank : 112) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 0.163984 (rank : 64) | NC score | 0.022073 (rank : 96) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
CSTFT_MOUSE
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.016343 (rank : 107) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 66) | NC score | 0.018548 (rank : 102) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 67) | NC score | 0.094061 (rank : 62) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 68) | NC score | 0.021812 (rank : 97) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
DNL3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 69) | NC score | 0.036404 (rank : 90) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 70) | NC score | 0.145279 (rank : 52) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 71) | NC score | 0.013903 (rank : 111) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ACM1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.003958 (rank : 143) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 908 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11229 | Gene names | CHRM1 | |||
|
Domain Architecture |
|
|||||
| Description | Muscarinic acetylcholine receptor M1. | |||||
|
ACTN2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.011867 (rank : 113) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
DNL3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 74) | NC score | 0.033468 (rank : 91) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 75) | NC score | 0.143529 (rank : 53) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 76) | NC score | 0.023922 (rank : 94) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 77) | NC score | 0.019266 (rank : 100) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.014608 (rank : 110) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
VIPR2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.006620 (rank : 131) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41588, P97750 | Gene names | Vipr2 | |||
|
Domain Architecture |

|
|||||
| Description | Vasoactive intestinal polypeptide receptor 2 precursor (VIP-R-2) (Pituitary adenylate cyclase-activating polypeptide type III receptor) (PACAP type III receptor) (PACAP-R-3). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | -0.002679 (rank : 162) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZN219_HUMAN
|
||||||
| θ value | 0.813845 (rank : 81) | NC score | 0.000842 (rank : 157) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
DJBP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.056103 (rank : 88) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.006358 (rank : 132) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
ACTN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.010780 (rank : 118) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.021006 (rank : 98) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.023749 (rank : 95) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.009603 (rank : 120) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
JHD2B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.007413 (rank : 128) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.070472 (rank : 79) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.005213 (rank : 137) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NFAC3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.008203 (rank : 125) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
TPTE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.090812 (rank : 64) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.003252 (rank : 146) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
DCNL4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.009743 (rank : 119) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92564, Q7Z3F3, Q7Z6B8 | Gene names | DCUN1D4, KIAA0276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCN1-like protein 4 (Defective in cullin neddylation protein 1-like protein 4) (DCUN1 domain-containing protein 4). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.018653 (rank : 101) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.027177 (rank : 93) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.009045 (rank : 121) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.017531 (rank : 105) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
ZN513_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | -0.004214 (rank : 165) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8E2 | Gene names | ZNF513 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 513. | |||||
|
CTND1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.003710 (rank : 145) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
GGEE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.008412 (rank : 124) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96GU1, Q8WWL9 | Gene names | PAGE5, GAGEE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G antigen family E member 1 (Prostate-associated gene 5 protein) (PAGE-5). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.006234 (rank : 134) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
ACTN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.008902 (rank : 122) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.008892 (rank : 123) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
FOXH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.002110 (rank : 153) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88621, Q9QZL5, Q9R241 | Gene names | Foxh1, Fast1, Fast2 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
GCNL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.010868 (rank : 116) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | -0.000136 (rank : 160) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.000140 (rank : 159) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
NPCL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.004810 (rank : 139) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6T3U4, Q5SVX1 | Gene names | Npc1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niemann-Pick C1-like protein 1 precursor. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.005930 (rank : 135) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.008153 (rank : 126) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.003077 (rank : 147) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.020817 (rank : 99) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TPTE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.129135 (rank : 57) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.002511 (rank : 150) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.016039 (rank : 108) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
ZDH18_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.002510 (rank : 151) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUE0, Q5JYH0, Q9H020 | Gene names | ZDHHC18 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC18 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 18) (DHHC-18). | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.006244 (rank : 133) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
DAB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.004376 (rank : 141) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |
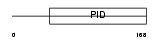
|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
DIPA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.015241 (rank : 109) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15834, Q96HA0 | Gene names | CCDC85B, DIPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-interacting protein A (Hepatitis delta antigen-interacting protein A) (Coiled-coil domain-containing protein 85B). | |||||
|
GLI3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | -0.004169 (rank : 164) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.001755 (rank : 154) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.065541 (rank : 83) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
NET2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.000473 (rank : 158) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00634 | Gene names | NTN2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor. | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.017049 (rank : 106) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | -0.003236 (rank : 163) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
ACM3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.000916 (rank : 156) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20309, Q4QRI3, Q5VXY2, Q9HB60 | Gene names | CHRM3 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M3. | |||||
|
APBB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.002884 (rank : 149) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
CSTFT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.005776 (rank : 136) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H0L4, O75174, Q53HK6, Q7LGE8, Q8N6T1 | Gene names | CSTF2T, KIAA0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
DTNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.001615 (rank : 155) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
IDD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.002934 (rank : 148) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |
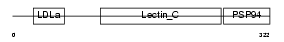
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
ITF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.004189 (rank : 142) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ITF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.003901 (rank : 144) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
JPH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.002345 (rank : 152) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
PARP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.006830 (rank : 130) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
SFR16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.010995 (rank : 115) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
SNIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.010809 (rank : 117) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TAD8, Q96SP9, Q9H9T7 | Gene names | SNIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.006993 (rank : 129) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
VASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.007635 (rank : 127) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
VSX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | -0.001804 (rank : 161) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
ZAR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.011839 (rank : 114) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.089795 (rank : 65) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.091824 (rank : 63) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.087338 (rank : 68) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.088323 (rank : 66) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.076768 (rank : 74) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.075844 (rank : 76) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.066669 (rank : 82) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.067064 (rank : 81) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.073798 (rank : 77) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.078688 (rank : 73) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.081435 (rank : 71) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.087756 (rank : 67) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.085412 (rank : 69) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.082549 (rank : 70) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.076457 (rank : 75) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.068665 (rank : 80) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.081209 (rank : 72) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.061205 (rank : 85) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.059889 (rank : 86) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.058787 (rank : 87) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
MCLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.061309 (rank : 84) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.100316 (rank : 61) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.072182 (rank : 78) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.051274 (rank : 89) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.997447 (rank : 2) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.993545 (rank : 3) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.993419 (rank : 4) | θ value | 0 (rank : 13) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.993397 (rank : 5) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.991918 (rank : 6) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.991536 (rank : 7) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.960243 (rank : 8) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.960135 (rank : 9) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.956055 (rank : 10) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.955464 (rank : 11) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.951368 (rank : 12) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.947736 (rank : 13) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.896416 (rank : 14) | θ value | 6.31527e-70 (rank : 16) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.870875 (rank : 15) | θ value | 3.58603e-65 (rank : 24) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.867686 (rank : 16) | θ value | 3.58603e-65 (rank : 23) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.859965 (rank : 17) | θ value | 1.00825e-67 (rank : 20) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.858226 (rank : 18) | θ value | 6.7473e-72 (rank : 14) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.852978 (rank : 19) | θ value | 1.15359e-63 (rank : 27) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.852519 (rank : 20) | θ value | 6.76295e-64 (rank : 26) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.850337 (rank : 21) | θ value | 4.52582e-68 (rank : 19) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.849894 (rank : 22) | θ value | 1.78386e-56 (rank : 30) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.843151 (rank : 23) | θ value | 1.83746e-69 (rank : 17) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.842081 (rank : 24) | θ value | 1.04338e-64 (rank : 25) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.840279 (rank : 25) | θ value | 4.83544e-70 (rank : 15) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.840048 (rank : 26) | θ value | 3.13423e-69 (rank : 18) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.839791 (rank : 27) | θ value | 6.53529e-67 (rank : 21) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.837717 (rank : 28) | θ value | 1.50663e-63 (rank : 28) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.837100 (rank : 29) | θ value | 3.24342e-66 (rank : 22) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.837087 (rank : 30) | θ value | 7.47731e-63 (rank : 29) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.392289 (rank : 31) | θ value | 3.64472e-09 (rank : 32) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.358209 (rank : 32) | θ value | 9.92553e-07 (rank : 35) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.355427 (rank : 33) | θ value | 5.81887e-07 (rank : 34) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.344946 (rank : 34) | θ value | 9.92553e-07 (rank : 36) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.339065 (rank : 35) | θ value | 1.17247e-07 (rank : 33) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.255439 (rank : 36) | θ value | 3.64472e-09 (rank : 31) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.240819 (rank : 37) | θ value | 0.000121331 (rank : 37) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.220106 (rank : 38) | θ value | 0.0148317 (rank : 47) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.214593 (rank : 39) | θ value | 0.00509761 (rank : 42) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.210964 (rank : 40) | θ value | 0.00509761 (rank : 41) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.207453 (rank : 41) | θ value | 0.0148317 (rank : 45) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.207381 (rank : 42) | θ value | 0.0148317 (rank : 46) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.191135 (rank : 43) | θ value | 0.00228821 (rank : 39) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.191010 (rank : 44) | θ value | 0.00228821 (rank : 38) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.187757 (rank : 45) | θ value | 0.00228821 (rank : 40) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.184874 (rank : 46) | θ value | 0.00665767 (rank : 44) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.184736 (rank : 47) | θ value | 0.00665767 (rank : 43) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.173619 (rank : 48) | θ value | 0.125558 (rank : 59) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.169855 (rank : 49) | θ value | 0.0431538 (rank : 54) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.161656 (rank : 50) | θ value | 0.0252991 (rank : 51) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.150767 (rank : 51) | θ value | 0.0148317 (rank : 48) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.145279 (rank : 52) | θ value | 0.62314 (rank : 70) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.143529 (rank : 53) | θ value | 0.813845 (rank : 75) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.141107 (rank : 54) | θ value | 0.0193708 (rank : 49) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.132833 (rank : 55) | θ value | 0.163984 (rank : 61) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.130541 (rank : 56) | θ value | 0.0252991 (rank : 50) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
TPTE2_HUMAN
|
||||||
| NC score | 0.129135 (rank : 57) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.128960 (rank : 58) | θ value | 0.0330416 (rank : 53) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.119194 (rank : 59) | θ value | 0.163984 (rank : 62) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.117896 (rank : 60) | θ value | 0.0736092 (rank : 56) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.100316 (rank : 61) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.094061 (rank : 62) | θ value | 0.365318 (rank : 67) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.091824 (rank : 63) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
TPTE_HUMAN
|
||||||
| NC score | 0.090812 (rank : 64) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.089795 (rank : 65) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.088323 (rank : 66) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.087756 (rank : 67) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.087338 (rank : 68) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.085412 (rank : 69) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.082549 (rank : 70) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.081435 (rank : 71) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.081209 (rank : 72) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.078688 (rank : 73) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.076768 (rank : 74) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.076457 (rank : 75) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.075844 (rank : 76) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.073798 (rank : 77) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.072182 (rank : 78) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.070472 (rank : 79) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.068665 (rank : 80) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.067064 (rank : 81) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.066669 (rank : 82) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.065541 (rank : 83) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
MCLN2_MOUSE
|
||||||
| NC score | 0.061309 (rank : 84) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.061205 (rank : 85) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.059889 (rank : 86) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.058787 (rank : 87) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
DJBP_MOUSE
|
||||||
| NC score | 0.056103 (rank : 88) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.051274 (rank : 89) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
DNL3_HUMAN
|
||||||
| NC score | 0.036404 (rank : 90) | θ value | 0.62314 (rank : 69) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL3_MOUSE
|
||||||
| NC score | 0.033468 (rank : 91) | θ value | 0.813845 (rank : 74) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.028378 (rank : 92) | θ value | 0.0736092 (rank : 55) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.027177 (rank : 93) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.023922 (rank : 94) | θ value | 0.813845 (rank : 76) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.023749 (rank : 95) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.022073 (rank : 96) | θ value | 0.163984 (rank : 64) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.021812 (rank : 97) | θ value | 0.365318 (rank : 68) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.021006 (rank : 98) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.020817 (rank : 99) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.019266 (rank : 100) | θ value | 0.813845 (rank : 77) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.018653 (rank : 101) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.018548 (rank : 102) | θ value | 0.365318 (rank : 66) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.018203 (rank : 103) | θ value | 0.0252991 (rank : 52) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
BSPRY_MOUSE
|
||||||
| NC score | 0.018101 (rank : 104) | θ value | 0.125558 (rank : 58) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YW5, Q3TU74, Q8BZF0, Q99KV7, Q9ER70 | Gene names | Bspry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B box and SPRY domain-containing protein. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.017531 (rank : 105) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.017049 (rank : 106) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
CSTFT_MOUSE
|
||||||
| NC score | 0.016343 (rank : 107) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.016039 (rank : 108) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
DIPA_HUMAN
|
||||||
| NC score | 0.015241 (rank : 109) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15834, Q96HA0 | Gene names | CCDC85B, DIPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-interacting protein A (Hepatitis delta antigen-interacting protein A) (Coiled-coil domain-containing protein 85B). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.014608 (rank : 110) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.013903 (rank : 111) | θ value | 0.62314 (rank : 71) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.013648 (rank : 112) | θ value | 0.163984 (rank : 63) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ACTN2_HUMAN
|
||||||
| NC score | 0.011867 (rank : 113) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ZAR1_HUMAN
|
||||||
| NC score | 0.011839 (rank : 114) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
SFR16_MOUSE
|
||||||
| NC score | 0.010995 (rank : 115) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
GCNL2_HUMAN
|
||||||
| NC score | 0.010868 (rank : 116) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
SNIP1_HUMAN
|
||||||
| NC score | 0.010809 (rank : 117) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TAD8, Q96SP9, Q9H9T7 | Gene names | SNIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
ACTN2_MOUSE
|
||||||
| NC score | 0.010780 (rank : 118) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
DCNL4_HUMAN
|
||||||
| NC score | 0.009743 (rank : 119) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92564, Q7Z3F3, Q7Z6B8 | Gene names | DCUN1D4, KIAA0276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCN1-like protein 4 (Defective in cullin neddylation protein 1-like protein 4) (DCUN1 domain-containing protein 4). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.009603 (rank : 120) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.009045 (rank : 121) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
ACTN3_HUMAN
|
||||||
| NC score | 0.008902 (rank : 122) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| NC score | 0.008892 (rank : 123) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
GGEE1_HUMAN
|
||||||
| NC score | 0.008412 (rank : 124) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96GU1, Q8WWL9 | Gene names | PAGE5, GAGEE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G antigen family E member 1 (Prostate-associated gene 5 protein) (PAGE-5). | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.008203 (rank : 125) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.008153 (rank : 126) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.007635 (rank : 127) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
JHD2B_MOUSE
|
||||||
| NC score | 0.007413 (rank : 128) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.006993 (rank : 129) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
PARP1_MOUSE
|
||||||
| NC score | 0.006830 (rank : 130) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
VIPR2_MOUSE
|
||||||
| NC score | 0.006620 (rank : 131) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41588, P97750 | Gene names | Vipr2 | |||
|
Domain Architecture |

|
|||||
| Description | Vasoactive intestinal polypeptide receptor 2 precursor (VIP-R-2) (Pituitary adenylate cyclase-activating polypeptide type III receptor) (PACAP type III receptor) (PACAP-R-3). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.006358 (rank : 132) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.006244 (rank : 133) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.006234 (rank : 134) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.005930 (rank : 135) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CSTFT_HUMAN
|
||||||
| NC score | 0.005776 (rank : 136) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H0L4, O75174, Q53HK6, Q7LGE8, Q8N6T1 | Gene names | CSTF2T, KIAA0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.005213 (rank : 137) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ACM5_HUMAN
|
||||||
| NC score | 0.004814 (rank : 138) | θ value | 0.125558 (rank : 57) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08912, Q96RG7 | Gene names | CHRM5 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M5. | |||||
|
NPCL1_MOUSE
|
||||||
| NC score | 0.004810 (rank : 139) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6T3U4, Q5SVX1 | Gene names | Npc1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niemann-Pick C1-like protein 1 precursor. | |||||
|
ACM1_MOUSE
|
||||||
| NC score | 0.004509 (rank : 140) | θ value | 0.163984 (rank : 60) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 917 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P12657, Q8BJN3 | Gene names | Chrm1, Chrm-1 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M1. | |||||
|
DAB2_MOUSE
|
||||||
| NC score | 0.004376 (rank : 141) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
ITF2_HUMAN
|
||||||
| NC score | 0.004189 (rank : 142) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ACM1_HUMAN
|
||||||
| NC score | 0.003958 (rank : 143) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 908 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11229 | Gene names | CHRM1 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M1. | |||||
|
ITF2_MOUSE
|
||||||
| NC score | 0.003901 (rank : 144) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
CTND1_HUMAN
|
||||||
| NC score | 0.003710 (rank : 145) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.003252 (rank : 146) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.003077 (rank : 147) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
IDD_HUMAN
|
||||||
| NC score | 0.002934 (rank : 148) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
APBB1_HUMAN
|
||||||
| NC score | 0.002884 (rank : 149) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.002511 (rank : 150) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
ZDH18_HUMAN
|
||||||
| NC score | 0.002510 (rank : 151) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUE0, Q5JYH0, Q9H020 | Gene names | ZDHHC18 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC18 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 18) (DHHC-18). | |||||
|
JPH1_MOUSE
|
||||||
| NC score | 0.002345 (rank : 152) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
FOXH1_MOUSE
|
||||||
| NC score | 0.002110 (rank : 153) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88621, Q9QZL5, Q9R241 | Gene names | Foxh1, Fast1, Fast2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.001755 (rank : 154) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
DTNA_HUMAN
|
||||||
| NC score | 0.001615 (rank : 155) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
ACM3_HUMAN
|
||||||
| NC score | 0.000916 (rank : 156) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20309, Q4QRI3, Q5VXY2, Q9HB60 | Gene names | CHRM3 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M3. | |||||
|
ZN219_HUMAN
|
||||||
| NC score | 0.000842 (rank : 157) | θ value | 0.813845 (rank : 81) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
NET2_HUMAN
|
||||||
| NC score | 0.000473 (rank : 158) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00634 | Gene names | NTN2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.000140 (rank : 159) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | -0.000136 (rank : 160) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
VSX1_MOUSE
|
||||||
| NC score | -0.001804 (rank : 161) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | -0.002679 (rank : 162) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZN447_HUMAN
|
||||||
| NC score | -0.003236 (rank : 163) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
GLI3_MOUSE
|
||||||
| NC score | -0.004169 (rank : 164) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
ZN513_HUMAN
|
||||||
| NC score | -0.004214 (rank : 165) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8E2 | Gene names | ZNF513 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 513. | |||||