Please be patient as the page loads
|
PARP1_MOUSE
|
||||||
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PARP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987324 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P09874, Q8IUZ9 | Gene names | PARP1, ADPRT, PPOL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1). | |||||
|
PARP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
PARP2_MOUSE
|
||||||
| θ value | 6.0463e-105 (rank : 3) | NC score | 0.907493 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88554, Q99N29 | Gene names | Parp2, Adprt2, Adprtl2, Aspartl2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (mPARP-2). | |||||
|
PARP2_HUMAN
|
||||||
| θ value | 2.80862e-102 (rank : 4) | NC score | 0.910424 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
PARP3_HUMAN
|
||||||
| θ value | 4.53632e-60 (rank : 5) | NC score | 0.876410 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 6) | NC score | 0.548923 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
DNL3_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 7) | NC score | 0.207811 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL3_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 8) | NC score | 0.205467 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 9) | NC score | 0.063699 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 10) | NC score | 0.067882 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.049046 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SPC25_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.084925 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HBM1 | Gene names | SPBC25, SPC25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25 (hSpc25). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.078719 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.033034 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
GLE1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.061557 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
PARP6_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.140392 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q2NL67, Q9H7C5, Q9H9X6, Q9HAF3, Q9NPS6, Q9UFG4 | Gene names | PARP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 6 (EC 2.4.2.30) (PARP-6). | |||||
|
PARP6_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.140328 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P6P7, Q8BVW1 | Gene names | Parp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 6 (EC 2.4.2.30) (PARP-6). | |||||
|
PARP8_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.140123 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N3A8, Q3KRB7, Q6DHZ1, Q9H754 | Gene names | PARP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 8 (EC 2.4.2.30) (PARP-8). | |||||
|
PARP8_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.139797 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3UD82, Q3UDE4, Q3UDU5, Q8VCB5, Q9CYY3 | Gene names | Parp8, D13Ertd275e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 8 (EC 2.4.2.30) (PARP-8). | |||||
|
KIF2C_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.021789 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99661, Q96C18, Q96HB8, Q9BWV8 | Gene names | KIF2C, KNSL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF2C (Mitotic centromere-associated kinesin) (MCAK) (Kinesin-like protein 6). | |||||
|
PEX14_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.061819 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0A0 | Gene names | Pex14 | |||
|
Domain Architecture |
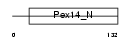
|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.036016 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.031201 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.069430 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
FA48A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.040842 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEM7, Q71RF3, Q9Y6A6 | Gene names | FAM48A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM48A (p38-interacting protein) (p38IP). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.014394 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
TOPB1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.043679 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92547, Q7LGC1, Q9UEB9 | Gene names | TOPBP1, KIAA0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase 2-binding protein 1 (DNA topoisomerase II-binding protein 1) (DNA topoisomerase IIbeta-binding protein 1) (TopBP1). | |||||
|
ISK5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.039680 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.031487 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.027912 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
BICD2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.034343 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.032105 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
FLNB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.022395 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75369, Q13706, Q8WXS9, Q8WXT0, Q8WXT1, Q8WXT2, Q9NRB5, Q9NT26, Q9UEV9 | Gene names | FLNB, FLN1L, FLN3, TABP, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP- 280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1). | |||||
|
THAP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.042041 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D305 | Gene names | Thap2 | |||
|
Domain Architecture |
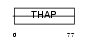
|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.030884 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.037349 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.030765 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.031926 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
PEX14_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.052411 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75381, Q8WX51 | Gene names | PEX14 | |||
|
Domain Architecture |
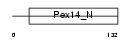
|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.031949 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.028752 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TTC5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.039004 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99LG4 | Gene names | Ttc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.024844 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.018144 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
DP13A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.025811 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
ENKUR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.035337 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.012861 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
DP13A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.024477 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
FLNB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.020835 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80X90, Q8VHX4, Q8VHX7, Q99KY3 | Gene names | Flnb | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (ABP- 280-like protein). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.016903 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.005717 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
FOXN4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.008441 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NZ1, Q6ZMR4, Q96NZ0 | Gene names | FOXN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein N4. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.024996 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.019533 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.023095 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PLCE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.019780 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUQ2, Q8IZ47, Q9BQG4 | Gene names | AGPAT5 | |||
|
Domain Architecture |
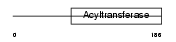
|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase epsilon (EC 2.3.1.51) (1-AGP acyltransferase 5) (1-AGPAT 5) (Lysophosphatidic acid acyltransferase-epsilon) (LPAAT-epsilon) (1-acylglycerol-3-phosphate O-acyltransferase 5). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.028533 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
STB5L_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.022761 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5DQR4, Q3TSM3, Q3UVG2, Q5DQR1, Q5DQR2, Q5DQR3, Q8BS52 | Gene names | Stxbp5l, Llgl4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5-like (Tomosyn-2) (Lethal(2) giant larvae protein homolog 4). | |||||
|
TTC5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.034669 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N0Z6 | Gene names | TTC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.020082 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.016393 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MMRN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.022619 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H8L6, Q6P2N2 | Gene names | MMRN2, EMILIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Multimerin-2 precursor (EMILIN-3) (Elastin microfibril interface located protein 3) (Elastin microfibril interfacer 3) (EndoGlyx-1 p125/p140 subunit). | |||||
|
MUSC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.012968 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.024727 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
P80C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.026657 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
PAR14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.045132 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
PARPT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.056935 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PARPT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.070769 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
RL7A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.027336 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12970, Q91YM3 | Gene names | Rpl7a, Surf-3, Surf3 | |||
|
Domain Architecture |
|
|||||
| Description | 60S ribosomal protein L7a (Surfeit locus protein 3). | |||||
|
STB5L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.022023 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2K9, Q4G1B4, Q6PIC3 | Gene names | STXBP5L, KIAA1006, LLGL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5-like (Tomosyn-2) (Lethal(2) giant larvae protein homolog 4). | |||||
|
XL3A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.033299 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60595 | Gene names | Xlr3a | |||
|
Domain Architecture |
|
|||||
| Description | X-linked lymphocyte-regulated protein 3A. | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.029347 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.010212 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.010194 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.025996 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.026008 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.021361 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DACH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.013933 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
K1196_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.007681 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
LYRIC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.016595 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WJ7, Q3U9F8, Q3UAQ8, Q8BN67, Q8CBT9, Q8CDL0, Q8CGI7, Q9D052 | Gene names | Mtdh, Lyric | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/LYRIC) (Metastasis adhesion protein) (Metadherin). | |||||
|
OFD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.035088 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.019705 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.030869 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
DEN2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.007219 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.013917 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.016120 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
RL7A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.023788 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62424, P11518 | Gene names | RPL7A, SURF-3, SURF3 | |||
|
Domain Architecture |
|
|||||
| Description | 60S ribosomal protein L7a (Surfeit locus protein 3) (PLA-X polypeptide). | |||||
|
TRFM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.009188 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08582, Q9BQE2 | Gene names | MFI2, MAP97 | |||
|
Domain Architecture |

|
|||||
| Description | Melanotransferrin precursor (Melanoma-associated antigen p97) (CD228 antigen). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.009995 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.006830 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.007271 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.021194 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CL011_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.011023 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NVM9, Q86WE2, Q96HM2, Q9BTX2, Q9NTB6, Q9NVM5 | Gene names | C12orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf11 (Sarcoma antigen NY-SAR-95). | |||||
|
ENKUR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.024463 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6SP97 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.008309 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
REST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.024951 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.023109 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.023651 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
STXB5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.019211 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T5C0, Q5T5C1, Q5T5C2, Q8NBG8, Q96NG9 | Gene names | STXBP5, LLGL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5 (Tomosyn-1) (Lethal(2) giant larvae protein homolog 3). | |||||
|
STXB5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.019317 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K400, Q80TM5, Q8K401, Q9D2F3 | Gene names | Stxbp5, Kiaa4253, Llgl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5 (Tomosyn-1) (Lethal(2) giant larvae protein homolog 3). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.022835 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.020379 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
LHR2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.051535 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
LHR2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.054855 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |
|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
PARP1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
PARP1_HUMAN
|
||||||
| NC score | 0.987324 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P09874, Q8IUZ9 | Gene names | PARP1, ADPRT, PPOL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1). | |||||
|
PARP2_HUMAN
|
||||||
| NC score | 0.910424 (rank : 3) | θ value | 2.80862e-102 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
PARP2_MOUSE
|
||||||
| NC score | 0.907493 (rank : 4) | θ value | 6.0463e-105 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88554, Q99N29 | Gene names | Parp2, Adprt2, Adprtl2, Aspartl2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (mPARP-2). | |||||
|
PARP3_HUMAN
|
||||||
| NC score | 0.876410 (rank : 5) | θ value | 4.53632e-60 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.548923 (rank : 6) | θ value | 4.43474e-23 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
DNL3_HUMAN
|
||||||
| NC score | 0.207811 (rank : 7) | θ value | 2.61198e-07 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL3_MOUSE
|
||||||
| NC score | 0.205467 (rank : 8) | θ value | 7.59969e-07 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
PARP6_HUMAN
|
||||||
| NC score | 0.140392 (rank : 9) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q2NL67, Q9H7C5, Q9H9X6, Q9HAF3, Q9NPS6, Q9UFG4 | Gene names | PARP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 6 (EC 2.4.2.30) (PARP-6). | |||||
|
PARP6_MOUSE
|
||||||
| NC score | 0.140328 (rank : 10) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P6P7, Q8BVW1 | Gene names | Parp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 6 (EC 2.4.2.30) (PARP-6). | |||||
|
PARP8_HUMAN
|
||||||
| NC score | 0.140123 (rank : 11) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N3A8, Q3KRB7, Q6DHZ1, Q9H754 | Gene names | PARP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 8 (EC 2.4.2.30) (PARP-8). | |||||
|
PARP8_MOUSE
|
||||||
| NC score | 0.139797 (rank : 12) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3UD82, Q3UDE4, Q3UDU5, Q8VCB5, Q9CYY3 | Gene names | Parp8, D13Ertd275e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 8 (EC 2.4.2.30) (PARP-8). | |||||
|
SPC25_HUMAN
|
||||||
| NC score | 0.084925 (rank : 13) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HBM1 | Gene names | SPBC25, SPC25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25 (hSpc25). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.078719 (rank : 14) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
PARPT_MOUSE
|
||||||
| NC score | 0.070769 (rank : 15) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.069430 (rank : 16) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.067882 (rank : 17) | θ value | 0.00869519 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.063699 (rank : 18) | θ value | 0.00665767 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
PEX14_MOUSE
|
||||||
| NC score | 0.061819 (rank : 19) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0A0 | Gene names | Pex14 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
GLE1_HUMAN
|
||||||
| NC score | 0.061557 (rank : 20) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
PARPT_HUMAN
|
||||||
| NC score | 0.056935 (rank : 21) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
LHR2A_MOUSE
|
||||||
| NC score | 0.054855 (rank : 22) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
PEX14_HUMAN
|
||||||
| NC score | 0.052411 (rank : 23) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75381, Q8WX51 | Gene names | PEX14 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
LHR2A_HUMAN
|
||||||
| NC score | 0.051535 (rank : 24) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.049046 (rank : 25) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
PAR14_MOUSE
|
||||||
| NC score | 0.045132 (rank : 26) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
TOPB1_HUMAN
|
||||||
| NC score | 0.043679 (rank : 27) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92547, Q7LGC1, Q9UEB9 | Gene names | TOPBP1, KIAA0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase 2-binding protein 1 (DNA topoisomerase II-binding protein 1) (DNA topoisomerase IIbeta-binding protein 1) (TopBP1). | |||||
|
THAP2_MOUSE
|
||||||
| NC score | 0.042041 (rank : 28) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D305 | Gene names | Thap2 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
FA48A_HUMAN
|
||||||
| NC score | 0.040842 (rank : 29) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEM7, Q71RF3, Q9Y6A6 | Gene names | FAM48A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM48A (p38-interacting protein) (p38IP). | |||||
|
ISK5_HUMAN
|
||||||
| NC score | 0.039680 (rank : 30) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
TTC5_MOUSE
|
||||||
| NC score | 0.039004 (rank : 31) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99LG4 | Gene names | Ttc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.037349 (rank : 32) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.036016 (rank : 33) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
ENKUR_HUMAN
|
||||||
| NC score | 0.035337 (rank : 34) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.035088 (rank : 35) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
TTC5_HUMAN
|
||||||
| NC score | 0.034669 (rank : 36) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N0Z6 | Gene names | TTC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
BICD2_HUMAN
|
||||||
| NC score | 0.034343 (rank : 37) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
XL3A_MOUSE
|
||||||
| NC score | 0.033299 (rank : 38) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60595 | Gene names | Xlr3a | |||
|
Domain Architecture |

|
|||||
| Description | X-linked lymphocyte-regulated protein 3A. | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.033034 (rank : 39) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.032105 (rank : 40) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.031949 (rank : 41) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.031926 (rank : 42) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.031487 (rank : 43) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.031201 (rank : 44) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.030884 (rank : 45) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.030869 (rank : 46) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.030765 (rank : 47) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.029347 (rank : 48) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.028752 (rank : 49) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.028533 (rank : 50) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.027912 (rank : 51) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
RL7A_MOUSE
|
||||||
| NC score | 0.027336 (rank : 52) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12970, Q91YM3 | Gene names | Rpl7a, Surf-3, Surf3 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L7a (Surfeit locus protein 3). | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.026657 (rank : 53) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.026008 (rank : 54) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.025996 (rank : 55) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DP13A_HUMAN
|
||||||
| NC score | 0.025811 (rank : 56) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.024996 (rank : 57) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.024951 (rank : 58) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.024844 (rank : 59) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.024727 (rank : 60) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.024477 (rank : 61) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
ENKUR_MOUSE
|
||||||
| NC score | 0.024463 (rank : 62) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6SP97 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
RL7A_HUMAN
|
||||||
| NC score | 0.023788 (rank : 63) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62424, P11518 | Gene names | RPL7A, SURF-3, SURF3 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L7a (Surfeit locus protein 3) (PLA-X polypeptide). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.023651 (rank : 64) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.023109 (rank : 65) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.023095 (rank : 66) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.022835 (rank : 67) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
STB5L_MOUSE
|
||||||
| NC score | 0.022761 (rank : 68) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5DQR4, Q3TSM3, Q3UVG2, Q5DQR1, Q5DQR2, Q5DQR3, Q8BS52 | Gene names | Stxbp5l, Llgl4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5-like (Tomosyn-2) (Lethal(2) giant larvae protein homolog 4). | |||||
|
MMRN2_HUMAN
|
||||||
| NC score | 0.022619 (rank : 69) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H8L6, Q6P2N2 | Gene names | MMRN2, EMILIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Multimerin-2 precursor (EMILIN-3) (Elastin microfibril interface located protein 3) (Elastin microfibril interfacer 3) (EndoGlyx-1 p125/p140 subunit). | |||||
|
FLNB_HUMAN
|
||||||
| NC score | 0.022395 (rank : 70) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75369, Q13706, Q8WXS9, Q8WXT0, Q8WXT1, Q8WXT2, Q9NRB5, Q9NT26, Q9UEV9 | Gene names | FLNB, FLN1L, FLN3, TABP, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP- 280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1). | |||||
|
STB5L_HUMAN
|
||||||
| NC score | 0.022023 (rank : 71) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2K9, Q4G1B4, Q6PIC3 | Gene names | STXBP5L, KIAA1006, LLGL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5-like (Tomosyn-2) (Lethal(2) giant larvae protein homolog 4). | |||||
|
KIF2C_HUMAN
|
||||||
| NC score | 0.021789 (rank : 72) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99661, Q96C18, Q96HB8, Q9BWV8 | Gene names | KIF2C, KNSL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF2C (Mitotic centromere-associated kinesin) (MCAK) (Kinesin-like protein 6). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.021361 (rank : 73) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.021194 (rank : 74) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
FLNB_MOUSE
|
||||||
| NC score | 0.020835 (rank : 75) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80X90, Q8VHX4, Q8VHX7, Q99KY3 | Gene names | Flnb | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (ABP- 280-like protein). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.020379 (rank : 76) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.020082 (rank : 77) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
PLCE_HUMAN
|
||||||
| NC score | 0.019780 (rank : 78) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUQ2, Q8IZ47, Q9BQG4 | Gene names | AGPAT5 | |||
|
Domain Architecture |

|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase epsilon (EC 2.3.1.51) (1-AGP acyltransferase 5) (1-AGPAT 5) (Lysophosphatidic acid acyltransferase-epsilon) (LPAAT-epsilon) (1-acylglycerol-3-phosphate O-acyltransferase 5). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.019705 (rank : 79) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.019533 (rank : 80) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
STXB5_MOUSE
|
||||||
| NC score | 0.019317 (rank : 81) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K400, Q80TM5, Q8K401, Q9D2F3 | Gene names | Stxbp5, Kiaa4253, Llgl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5 (Tomosyn-1) (Lethal(2) giant larvae protein homolog 3). | |||||
|
STXB5_HUMAN
|
||||||
| NC score | 0.019211 (rank : 82) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T5C0, Q5T5C1, Q5T5C2, Q8NBG8, Q96NG9 | Gene names | STXBP5, LLGL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5 (Tomosyn-1) (Lethal(2) giant larvae protein homolog 3). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.018144 (rank : 83) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.016903 (rank : 84) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
LYRIC_MOUSE
|
||||||
| NC score | 0.016595 (rank : 85) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WJ7, Q3U9F8, Q3UAQ8, Q8BN67, Q8CBT9, Q8CDL0, Q8CGI7, Q9D052 | Gene names | Mtdh, Lyric | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/LYRIC) (Metastasis adhesion protein) (Metadherin). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.016393 (rank : 86) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.016120 (rank : 87) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.014394 (rank : 88) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
DACH1_HUMAN
|
||||||
| NC score | 0.013933 (rank : 89) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.013917 (rank : 90) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
MUSC_HUMAN
|
||||||
| NC score | 0.012968 (rank : 91) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.012861 (rank : 92) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CL011_HUMAN
|
||||||
| NC score | 0.011023 (rank : 93) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NVM9, Q86WE2, Q96HM2, Q9BTX2, Q9NTB6, Q9NVM5 | Gene names | C12orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf11 (Sarcoma antigen NY-SAR-95). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.010212 (rank : 94) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.010194 (rank : 95) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.009995 (rank : 96) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TRFM_HUMAN
|
||||||
| NC score | 0.009188 (rank : 97) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08582, Q9BQE2 | Gene names | MFI2, MAP97 | |||
|
Domain Architecture |

|
|||||
| Description | Melanotransferrin precursor (Melanoma-associated antigen p97) (CD228 antigen). | |||||
|
FOXN4_HUMAN
|
||||||
| NC score | 0.008441 (rank : 98) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NZ1, Q6ZMR4, Q96NZ0 | Gene names | FOXN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein N4. | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.008309 (rank : 99) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
K1196_HUMAN
|
||||||
| NC score | 0.007681 (rank : 100) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.007271 (rank : 101) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
DEN2A_MOUSE
|
||||||
| NC score | 0.007219 (rank : 102) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.006830 (rank : 103) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.005717 (rank : 104) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||