Please be patient as the page loads
|
ENKUR_HUMAN
|
||||||
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ENKUR_HUMAN
|
||||||
| θ value | 2.689e-137 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
ENKUR_MOUSE
|
||||||
| θ value | 4.45295e-124 (rank : 2) | NC score | 0.972155 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6SP97 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 3) | NC score | 0.127032 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
GEMI_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 4) | NC score | 0.168928 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75496, Q9H1Z1 | Gene names | GMNN | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
GEMI_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.191983 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
TRIM6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 6) | NC score | 0.044162 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9C030, Q86WZ8, Q9HCR1 | Gene names | TRIM6, RNF89 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 6 (RING finger protein 89). | |||||
|
TRIM6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.046311 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BGE7, Q99PQ6 | Gene names | Trim6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 6. | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 0.47712 (rank : 8) | NC score | 0.020520 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.036662 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.036199 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.036101 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.037776 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
TRI60_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.032885 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q495X7, Q8NA35 | Gene names | TRIM60, RNF129 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 60 (RING finger protein 129). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.045524 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.043936 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
K1377_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.063071 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2H0, Q4G0U6 | Gene names | KIAA1377 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1377. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.025666 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.042696 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.040164 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
RO60_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.069322 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08848, Q9QYD8 | Gene names | Trove2, Ssa2 | |||
|
Domain Architecture |

|
|||||
| Description | 60 kDa SS-A/Ro ribonucleoprotein (60 kDa Ro protein) (60 kDa ribonucleoprotein Ro) (RoRNP) (TROVE domain family member 2). | |||||
|
CAD17_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.013603 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12864, Q15336 | Gene names | CDH17 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-17 precursor (Liver-intestine-cadherin) (LI-cadherin) (Intestinal peptide-associated transporter HPT-1). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.045698 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.036664 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.012975 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TTK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.010433 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35761 | Gene names | Ttk, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (ESK) (PYT). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.042316 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
PARP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.035337 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
PHF2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.021515 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.040751 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
SEPT6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.021789 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R1T4, Q91XH2, Q9CZ94 | Gene names | Sept6 | |||
|
Domain Architecture |
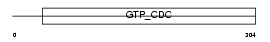
|
|||||
| Description | Septin-6. | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.037099 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.035087 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.034667 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.039092 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.030856 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
MPP10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.037079 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
MYO6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.035033 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
NKX31_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.007667 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97436, O09087 | Gene names | Nkx3-1, Nkx-3.1, Nkx3a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-3.1. | |||||
|
REPS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.028323 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.027187 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CEP55_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.037345 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
CS018_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.096412 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEA5 | Gene names | C19orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf18 precursor. | |||||
|
DNJ5G_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.015756 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
K0831_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.031754 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
KAD7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.026154 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96M32, Q8IYP6 | Gene names | AK7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
SEPT6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.019525 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14141, Q5JTK0, Q969W5, Q96A13, Q96GR1, Q96P86, Q96P87 | Gene names | SEPT6, KIAA0128, SEP2 | |||
|
Domain Architecture |
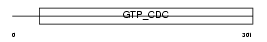
|
|||||
| Description | Septin-6. | |||||
|
CKAP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.025288 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14008, Q14668 | Gene names | CKAP5, KIAA0097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 5 (Colonic and hepatic tumor over- expressed protein) (Ch-TOG protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.037101 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.080419 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
SPFH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.027380 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91X78, Q8CIH7 | Gene names | Spfh1, Keo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPFH domain-containing protein 1 precursor (KE04 protein homolog). | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.035104 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
UBR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.014590 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWV7, O60708, O75492, Q68DN9, Q8IWY6, Q96JY4 | Gene names | UBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
BIN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.021877 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQY0, Q9BVG2, Q9NVY9 | Gene names | BIN3 | |||
|
Domain Architecture |
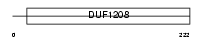
|
|||||
| Description | Bridging integrator 3. | |||||
|
CASZ1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.014677 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.023517 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CHM4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.016183 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BY43, Q32Q79, Q86SZ8, Q96QJ9, Q9P026 | Gene names | CHMP4A, C14orf123, SHAX2 | |||
|
Domain Architecture |
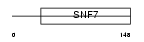
|
|||||
| Description | Charged multivesicular body protein 4a (Chromatin-modifying protein 4a) (CHMP4a) (Vacuolar protein sorting 7-1) (SNF7-1) (hSnf-1) (SNF7 homolog associated with Alix-2). | |||||
|
CM007_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.028886 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5W0B1, Q8TBY2, Q9H0T2, Q9H8M0 | Gene names | C13orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7. | |||||
|
DX26B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.025896 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JSJ4, Q5CZA2, Q6IPS3, Q6ZTU5, Q6ZWE4 | Gene names | DDX26B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
DX26B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.025798 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BND4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
EI2BD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.015605 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61749, Q61748, Q8VC35 | Gene names | Eif2b4, Eif2b, Eif2bd, Jgr1a | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
|
H14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.020532 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
K0020_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.018587 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15397, Q547G7, Q5SZY9, Q6IB47, Q96B27, Q96L78, Q96L79, Q96L80 | Gene names | KIAA0020, XTP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0020 (HBV X-transactivated gene 5 protein) (Minor histocompatibility antigen HA-8) (HLA-HA8). | |||||
|
M3K5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.004798 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35099 | Gene names | Map3k5, Ask1, Mekk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 5 (EC 2.7.11.25) (MAPK/ERK kinase kinase 5) (MEK kinase 5) (MEKK 5) (Apoptosis signal- regulating kinase 1) (ASK-1). | |||||
|
TFAM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.037566 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.055466 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ENKUR_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.689e-137 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
ENKUR_MOUSE
|
||||||
| NC score | 0.972155 (rank : 2) | θ value | 4.45295e-124 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6SP97 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
GEMI_MOUSE
|
||||||
| NC score | 0.191983 (rank : 3) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
GEMI_HUMAN
|
||||||
| NC score | 0.168928 (rank : 4) | θ value | 0.0563607 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75496, Q9H1Z1 | Gene names | GMNN | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.127032 (rank : 5) | θ value | 7.1131e-05 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
CS018_HUMAN
|
||||||
| NC score | 0.096412 (rank : 6) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEA5 | Gene names | C19orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf18 precursor. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.080419 (rank : 7) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RO60_MOUSE
|
||||||
| NC score | 0.069322 (rank : 8) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08848, Q9QYD8 | Gene names | Trove2, Ssa2 | |||
|
Domain Architecture |

|
|||||
| Description | 60 kDa SS-A/Ro ribonucleoprotein (60 kDa Ro protein) (60 kDa ribonucleoprotein Ro) (RoRNP) (TROVE domain family member 2). | |||||
|
K1377_HUMAN
|
||||||
| NC score | 0.063071 (rank : 9) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2H0, Q4G0U6 | Gene names | KIAA1377 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1377. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.055466 (rank : 10) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TRIM6_MOUSE
|
||||||
| NC score | 0.046311 (rank : 11) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BGE7, Q99PQ6 | Gene names | Trim6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 6. | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.045698 (rank : 12) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.045524 (rank : 13) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
TRIM6_HUMAN
|
||||||
| NC score | 0.044162 (rank : 14) | θ value | 0.125558 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9C030, Q86WZ8, Q9HCR1 | Gene names | TRIM6, RNF89 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 6 (RING finger protein 89). | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.043936 (rank : 15) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.042696 (rank : 16) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.042316 (rank : 17) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.040751 (rank : 18) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.040164 (rank : 19) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.039092 (rank : 20) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.037776 (rank : 21) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
TFAM_HUMAN
|
||||||
| NC score | 0.037566 (rank : 22) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.037345 (rank : 23) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.037101 (rank : 24) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.037099 (rank : 25) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
MPP10_HUMAN
|
||||||
| NC score | 0.037079 (rank : 26) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.036664 (rank : 27) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.036662 (rank : 28) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.036199 (rank : 29) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.036101 (rank : 30) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
PARP1_MOUSE
|
||||||
| NC score | 0.035337 (rank : 31) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.035104 (rank : 32) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.035087 (rank : 33) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
MYO6_MOUSE
|
||||||
| NC score | 0.035033 (rank : 34) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.034667 (rank : 35) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
TRI60_HUMAN
|
||||||
| NC score | 0.032885 (rank : 36) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q495X7, Q8NA35 | Gene names | TRIM60, RNF129 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 60 (RING finger protein 129). | |||||
|
K0831_MOUSE
|
||||||
| NC score | 0.031754 (rank : 37) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.030856 (rank : 38) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CM007_HUMAN
|
||||||
| NC score | 0.028886 (rank : 39) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5W0B1, Q8TBY2, Q9H0T2, Q9H8M0 | Gene names | C13orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7. | |||||
|
REPS2_HUMAN
|
||||||
| NC score | 0.028323 (rank : 40) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
SPFH1_MOUSE
|
||||||
| NC score | 0.027380 (rank : 41) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91X78, Q8CIH7 | Gene names | Spfh1, Keo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPFH domain-containing protein 1 precursor (KE04 protein homolog). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.027187 (rank : 42) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
KAD7_HUMAN
|
||||||
| NC score | 0.026154 (rank : 43) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96M32, Q8IYP6 | Gene names | AK7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
DX26B_HUMAN
|
||||||
| NC score | 0.025896 (rank : 44) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JSJ4, Q5CZA2, Q6IPS3, Q6ZTU5, Q6ZWE4 | Gene names | DDX26B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
DX26B_MOUSE
|
||||||
| NC score | 0.025798 (rank : 45) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BND4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.025666 (rank : 46) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CKAP5_HUMAN
|
||||||
| NC score | 0.025288 (rank : 47) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14008, Q14668 | Gene names | CKAP5, KIAA0097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 5 (Colonic and hepatic tumor over- expressed protein) (Ch-TOG protein). | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.023517 (rank : 48) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
BIN3_HUMAN
|
||||||
| NC score | 0.021877 (rank : 49) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQY0, Q9BVG2, Q9NVY9 | Gene names | BIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Bridging integrator 3. | |||||
|
SEPT6_MOUSE
|
||||||
| NC score | 0.021789 (rank : 50) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R1T4, Q91XH2, Q9CZ94 | Gene names | Sept6 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-6. | |||||
|
PHF2_HUMAN
|
||||||
| NC score | 0.021515 (rank : 51) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.020532 (rank : 52) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.020520 (rank : 53) | θ value | 0.47712 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
SEPT6_HUMAN
|
||||||
| NC score | 0.019525 (rank : 54) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14141, Q5JTK0, Q969W5, Q96A13, Q96GR1, Q96P86, Q96P87 | Gene names | SEPT6, KIAA0128, SEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-6. | |||||
|
K0020_HUMAN
|
||||||
| NC score | 0.018587 (rank : 55) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15397, Q547G7, Q5SZY9, Q6IB47, Q96B27, Q96L78, Q96L79, Q96L80 | Gene names | KIAA0020, XTP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0020 (HBV X-transactivated gene 5 protein) (Minor histocompatibility antigen HA-8) (HLA-HA8). | |||||
|
CHM4A_HUMAN
|
||||||
| NC score | 0.016183 (rank : 56) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BY43, Q32Q79, Q86SZ8, Q96QJ9, Q9P026 | Gene names | CHMP4A, C14orf123, SHAX2 | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4a (Chromatin-modifying protein 4a) (CHMP4a) (Vacuolar protein sorting 7-1) (SNF7-1) (hSnf-1) (SNF7 homolog associated with Alix-2). | |||||
|
DNJ5G_HUMAN
|
||||||
| NC score | 0.015756 (rank : 57) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
EI2BD_MOUSE
|
||||||
| NC score | 0.015605 (rank : 58) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61749, Q61748, Q8VC35 | Gene names | Eif2b4, Eif2b, Eif2bd, Jgr1a | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
|
CASZ1_HUMAN
|
||||||
| NC score | 0.014677 (rank : 59) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
UBR1_HUMAN
|
||||||
| NC score | 0.014590 (rank : 60) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWV7, O60708, O75492, Q68DN9, Q8IWY6, Q96JY4 | Gene names | UBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
CAD17_HUMAN
|
||||||
| NC score | 0.013603 (rank : 61) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12864, Q15336 | Gene names | CDH17 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-17 precursor (Liver-intestine-cadherin) (LI-cadherin) (Intestinal peptide-associated transporter HPT-1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.012975 (rank : 62) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TTK_MOUSE
|
||||||
| NC score | 0.010433 (rank : 63) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35761 | Gene names | Ttk, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (ESK) (PYT). | |||||
|
NKX31_MOUSE
|
||||||
| NC score | 0.007667 (rank : 64) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97436, O09087 | Gene names | Nkx3-1, Nkx-3.1, Nkx3a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-3.1. | |||||
|
M3K5_MOUSE
|
||||||
| NC score | 0.004798 (rank : 65) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35099 | Gene names | Map3k5, Ask1, Mekk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 5 (EC 2.7.11.25) (MAPK/ERK kinase kinase 5) (MEK kinase 5) (MEKK 5) (Apoptosis signal- regulating kinase 1) (ASK-1). | |||||