Please be patient as the page loads
|
KAD7_HUMAN
|
||||||
| SwissProt Accessions | Q96M32, Q8IYP6 | Gene names | AK7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KAD7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q96M32, Q8IYP6 | Gene names | AK7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
KAD7_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982719 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D2H2, Q8BVH3 | Gene names | Ak7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
DPY30_MOUSE
|
||||||
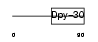
| θ value | 2.61198e-07 (rank : 3) | NC score | 0.501158 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99LT0 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Dpy-30-like protein. | |||||
|
DPY30_HUMAN
|
||||||
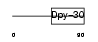
| θ value | 3.41135e-07 (rank : 4) | NC score | 0.497976 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C005 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Dpy-30-like protein. | |||||
|
NDK5_HUMAN
|
||||||
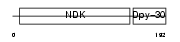
| θ value | 2.21117e-06 (rank : 5) | NC score | 0.261180 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P56597 | Gene names | NME5 | |||
|
Domain Architecture |
|
|||||
| Description | Nucleoside diphosphate kinase homolog 5 (NDK-H 5) (NDP kinase homolog 5) (nm23-H5) (Testis-specific nm23 homolog) (Inhibitor of p53-induced apoptosis-beta) (IPIA-beta). | |||||
|
NDK5_MOUSE
|
||||||
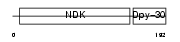
| θ value | 3.77169e-06 (rank : 6) | NC score | 0.260320 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MH5, Q810R1 | Gene names | Nme5 | |||
|
Domain Architecture |
|
|||||
| Description | Nucleoside diphosphate kinase homolog 5 (NDK-H 5) (NDP kinase homolog 5) (nm23-M5). | |||||
|
KAD2_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 7) | NC score | 0.280520 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTP6, Q3TKI6, Q9CY37 | Gene names | Ak2, Ak-2 | |||
|
Domain Architecture |
|
|||||
| Description | Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
KAD2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 8) | NC score | 0.274406 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54819, Q16856, Q5TIF7, Q8TCY2, Q8TCY3 | Gene names | AK2, ADK2 | |||
|
Domain Architecture |
|
|||||
| Description | Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
KAD4_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 9) | NC score | 0.235034 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27144 | Gene names | AK3L1, AK3, AK4 | |||
|
Domain Architecture |
|
|||||
| Description | Adenylate kinase isoenzyme 4, mitochondrial (EC 2.7.4.3) (Adenylate kinase 3-like 1) (ATP-AMP transphosphorylase). | |||||
|
KAD4_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 10) | NC score | 0.222068 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUR9, Q9R1X7 | Gene names | Ak3l1, Ak-4, Ak3b, Ak4 | |||
|
Domain Architecture |
|
|||||
| Description | Adenylate kinase isoenzyme 4, mitochondrial (EC 2.7.4.3) (Adenylate kinase 3-like 1) (ATP-AMP transphosphorylase). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 11) | NC score | 0.109171 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 12) | NC score | 0.107641 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.059006 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.064400 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
DYDC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.181953 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WWB3, Q5QP03, Q5QP04, Q6WNP4, Q6ZU87 | Gene names | DYDC1, DPY30D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 1 (RSD-9). | |||||
|
DYDC1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.203189 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D9T0, Q810Q6 | Gene names | Dydc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 1. | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.049374 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
EVPL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.055486 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
CDC37_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.063633 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
KAD3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.187742 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WTP7, Q3UDN7, Q8BGX5, Q9D7Z1, Q9DB57, Q9DBM5 | Gene names | Ak3, Ak3l, Ak3l1, Akl3l | |||
|
Domain Architecture |
|
|||||
| Description | GTP:AMP phosphotransferase mitochondrial (EC 2.7.4.10) (Adenylate kinase 3) (AK3) (Adenylate kinase 3 alpha-like 1). | |||||
|
STX4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.027914 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70452 | Gene names | Stx4, Stx4a | |||
|
Domain Architecture |
|
|||||
| Description | Syntaxin-4. | |||||
|
DYDC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.146853 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96IM9, Q5QP07, Q5QP11 | Gene names | DYDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 2. | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.057398 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.057408 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
ITPR2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.026238 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
TRPM6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.023089 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.052888 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.043119 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.040763 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
GEMI_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.073828 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75496, Q9H1Z1 | Gene names | GMNN | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
KAD5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.121785 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q920P5 | Gene names | Ak5 | |||
|
Domain Architecture |
|
|||||
| Description | Adenylate kinase isoenzyme 5 (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
MERL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.036867 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.036906 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.046236 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.046415 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
THNSL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.034356 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.043446 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.039580 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
GEMI_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.075179 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
IF2M_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.027762 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
MERL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.036437 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.047647 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.039457 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.044543 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CCDC5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.038949 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96CS2, Q8N837 | Gene names | CCDC5, HEIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 5 (Enhancer of invasion-cluster) (HEI-C). | |||||
|
CLH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.036381 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00610, Q6N0A0, Q86TF2 | Gene names | CLTC, CLH17, KIAA0034 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 1 (CLH-17). | |||||
|
CLH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.036376 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68FD5 | Gene names | Cltc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin heavy chain. | |||||
|
CT077_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.045521 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQG5 | Gene names | C20orf77 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf77. | |||||
|
KAD3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.152446 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UIJ7, Q5VYW6, Q9H576, Q9NPB4 | Gene names | AK3, AK3L1, AKL3L | |||
|
Domain Architecture |
|
|||||
| Description | GTP:AMP phosphotransferase mitochondrial (EC 2.7.4.10) (Adenylate kinase 3) (AK3) (Adenylate kinase 3 alpha-like 1). | |||||
|
KI13B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.016983 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.058859 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
LRC48_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.018753 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H069, Q86SF9, Q86W73, Q8IWG0 | Gene names | LRRC48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 48. | |||||
|
RGNEF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.024804 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.025589 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.025584 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
AR13B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.020780 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
ATAD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.024099 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.024099 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.030803 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
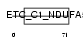
NDUA5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.059002 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CPP6, Q9CY90, Q9D2P2, Q9D703, Q9D739 | Gene names | Ndufa5 | |||
|
Domain Architecture |
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 13 kDa-B subunit) (Complex I-13kD-B) (CI-13kD-B) (Complex I subunit B13). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.038621 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.027147 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
ALAT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.015165 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGT5, Q8BVY7, Q8K1J3 | Gene names | Gpt2, Aat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alanine aminotransferase 2 (EC 2.6.1.2) (ALT2) (Glutamic--pyruvic transaminase 2) (GPT 2) (Glutamic--alanine transaminase 2). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.032171 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
ENKUR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.026154 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.033297 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.011285 (rank : 92) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
ABCG2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.007758 (rank : 94) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TMS5, Q9R004, Q9Z1T0 | Gene names | Abcg2, Abcp, Bcrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family G member 2 (Breast cancer resistance protein 1 homolog). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.025398 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
DDX56_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.012092 (rank : 91) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.015214 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.023230 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
KAD6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.044925 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
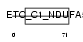
NDUA5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.048980 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16718, Q6IRX7 | Gene names | NDUFA5 | |||
|
Domain Architecture |
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 13 kDa-B subunit) (Complex I-13kD-B) (CI-13kD-B) (Complex I subunit B13). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.014602 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
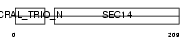
CT121_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.010772 (rank : 93) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTX7, Q5QPC1, Q9H1G2, Q9NQG8 | Gene names | C20orf121 | |||
|
Domain Architecture |
|
|||||
| Description | Protein C20orf121. | |||||
|
ITPR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.020293 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.031388 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.031113 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.053944 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.014273 (rank : 90) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.022178 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.022179 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
KAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.097605 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P00568, Q9BVK9, Q9UQC7 | Gene names | AK1 | |||
|
Domain Architecture |
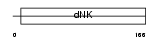
|
|||||
| Description | Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase). | |||||
|
KAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.093961 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0Y5 | Gene names | Ak1 | |||
|
Domain Architecture |
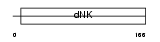
|
|||||
| Description | Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase). | |||||
|
KAD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.087605 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6K8, Q96EC9 | Gene names | AK5 | |||
|
Domain Architecture |
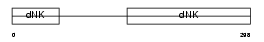
|
|||||
| Description | Adenylate kinase isoenzyme 5 (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
KCY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.086647 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P30085, Q53GB7, Q5SVZ0, Q96C07, Q9UBQ8, Q9UIA2 | Gene names | CMPK, CMK, UCK, UMK, UMPK | |||
|
Domain Architecture |
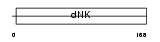
|
|||||
| Description | UMP-CMP kinase (EC 2.7.4.14) (Cytidylate kinase) (Deoxycytidylate kinase) (Cytidine monophosphate kinase) (Uridine monophosphate/cytidine monophosphate kinase) (UMP/CMP kinase) (UMP/CMPK) (Uridine monophosphate kinase). | |||||
|
KCY_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.086937 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBP5 | Gene names | Cmpk, Cmk, Uck, Umk, Umpk | |||
|
Domain Architecture |
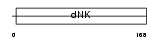
|
|||||
| Description | UMP-CMP kinase (EC 2.7.4.14) (Cytidylate kinase) (Deoxycytidylate kinase) (Cytidine monophosphate kinase) (Uridine monophosphate/cytidine monophosphate kinase) (UMP/CMP kinase) (UMP/CMPK) (Uridine monophosphate kinase). | |||||
|
NDK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.051715 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75414, Q96E73, Q9BQ63 | Gene names | NME6 | |||
|
Domain Architecture |
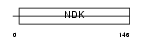
|
|||||
| Description | Nucleoside diphosphate kinase 6 (EC 2.7.4.6) (NDK 6) (NDP kinase 6) (nm23-H6) (Inhibitor of p53-induced apoptosis-alpha) (IPIA-alpha). | |||||
|
NDK6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.051708 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88425, Q99M53 | Gene names | Nme6 | |||
|
Domain Architecture |
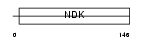
|
|||||
| Description | Nucleoside diphosphate kinase 6 (EC 2.7.4.6) (NDK 6) (NDP kinase 6) (nm23-M6). | |||||
|
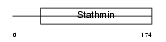
STMN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.052508 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q93045, O14952, Q6PK68 | Gene names | STMN2, SCG10, SCGN10 | |||
|
Domain Architecture |
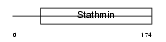
|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
STMN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.052508 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P55821 | Gene names | Stmn2, Scg10, Scgn10, Stmb2 | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
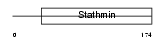
STMN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.050005 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZ72, O75527, Q969Y4 | Gene names | STMN3, SCLIP | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-3 (SCG10-like protein). | |||||
|
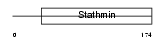
STMN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.050390 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O70166 | Gene names | Stmn3, Sclip | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-3 (SCG10-like protein) (SCG10-related protein HiAT3) (Hippocampus abundant transcript 3). | |||||
|
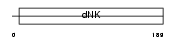
KAD7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q96M32, Q8IYP6 | Gene names | AK7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
KAD7_MOUSE
|
||||||
| NC score | 0.982719 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D2H2, Q8BVH3 | Gene names | Ak7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
DPY30_MOUSE
|
||||||
| NC score | 0.501158 (rank : 3) | θ value | 2.61198e-07 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99LT0 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Dpy-30-like protein. | |||||
|
DPY30_HUMAN
|
||||||
| NC score | 0.497976 (rank : 4) | θ value | 3.41135e-07 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C005 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Dpy-30-like protein. | |||||
|
KAD2_MOUSE
|
||||||
| NC score | 0.280520 (rank : 5) | θ value | 1.09739e-05 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTP6, Q3TKI6, Q9CY37 | Gene names | Ak2, Ak-2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
KAD2_HUMAN
|
||||||
| NC score | 0.274406 (rank : 6) | θ value | 2.44474e-05 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54819, Q16856, Q5TIF7, Q8TCY2, Q8TCY3 | Gene names | AK2, ADK2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
NDK5_HUMAN
|
||||||
| NC score | 0.261180 (rank : 7) | θ value | 2.21117e-06 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P56597 | Gene names | NME5 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoside diphosphate kinase homolog 5 (NDK-H 5) (NDP kinase homolog 5) (nm23-H5) (Testis-specific nm23 homolog) (Inhibitor of p53-induced apoptosis-beta) (IPIA-beta). | |||||
|
NDK5_MOUSE
|
||||||
| NC score | 0.260320 (rank : 8) | θ value | 3.77169e-06 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MH5, Q810R1 | Gene names | Nme5 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoside diphosphate kinase homolog 5 (NDK-H 5) (NDP kinase homolog 5) (nm23-M5). | |||||
|
KAD4_HUMAN
|
||||||
| NC score | 0.235034 (rank : 9) | θ value | 0.00035302 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27144 | Gene names | AK3L1, AK3, AK4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 4, mitochondrial (EC 2.7.4.3) (Adenylate kinase 3-like 1) (ATP-AMP transphosphorylase). | |||||
|
KAD4_MOUSE
|
||||||
| NC score | 0.222068 (rank : 10) | θ value | 0.00228821 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUR9, Q9R1X7 | Gene names | Ak3l1, Ak-4, Ak3b, Ak4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 4, mitochondrial (EC 2.7.4.3) (Adenylate kinase 3-like 1) (ATP-AMP transphosphorylase). | |||||
|
DYDC1_MOUSE
|
||||||
| NC score | 0.203189 (rank : 11) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D9T0, Q810Q6 | Gene names | Dydc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 1. | |||||
|
KAD3_MOUSE
|
||||||
| NC score | 0.187742 (rank : 12) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WTP7, Q3UDN7, Q8BGX5, Q9D7Z1, Q9DB57, Q9DBM5 | Gene names | Ak3, Ak3l, Ak3l1, Akl3l | |||
|
Domain Architecture |

|
|||||
| Description | GTP:AMP phosphotransferase mitochondrial (EC 2.7.4.10) (Adenylate kinase 3) (AK3) (Adenylate kinase 3 alpha-like 1). | |||||
|
DYDC1_HUMAN
|
||||||
| NC score | 0.181953 (rank : 13) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WWB3, Q5QP03, Q5QP04, Q6WNP4, Q6ZU87 | Gene names | DYDC1, DPY30D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 1 (RSD-9). | |||||
|
KAD3_HUMAN
|
||||||
| NC score | 0.152446 (rank : 14) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UIJ7, Q5VYW6, Q9H576, Q9NPB4 | Gene names | AK3, AK3L1, AKL3L | |||
|
Domain Architecture |

|
|||||
| Description | GTP:AMP phosphotransferase mitochondrial (EC 2.7.4.10) (Adenylate kinase 3) (AK3) (Adenylate kinase 3 alpha-like 1). | |||||
|
DYDC2_HUMAN
|
||||||
| NC score | 0.146853 (rank : 15) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96IM9, Q5QP07, Q5QP11 | Gene names | DYDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 2. | |||||
|
KAD5_MOUSE
|
||||||
| NC score | 0.121785 (rank : 16) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q920P5 | Gene names | Ak5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 5 (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.109171 (rank : 17) | θ value | 0.00298849 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.107641 (rank : 18) | θ value | 0.00390308 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
KAD1_HUMAN
|
||||||
| NC score | 0.097605 (rank : 19) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P00568, Q9BVK9, Q9UQC7 | Gene names | AK1 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase). | |||||
|
KAD1_MOUSE
|
||||||
| NC score | 0.093961 (rank : 20) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0Y5 | Gene names | Ak1 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase). | |||||
|
KAD5_HUMAN
|
||||||
| NC score | 0.087605 (rank : 21) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6K8, Q96EC9 | Gene names | AK5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 5 (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
KCY_MOUSE
|
||||||
| NC score | 0.086937 (rank : 22) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBP5 | Gene names | Cmpk, Cmk, Uck, Umk, Umpk | |||
|
Domain Architecture |

|
|||||
| Description | UMP-CMP kinase (EC 2.7.4.14) (Cytidylate kinase) (Deoxycytidylate kinase) (Cytidine monophosphate kinase) (Uridine monophosphate/cytidine monophosphate kinase) (UMP/CMP kinase) (UMP/CMPK) (Uridine monophosphate kinase). | |||||
|
KCY_HUMAN
|
||||||
| NC score | 0.086647 (rank : 23) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P30085, Q53GB7, Q5SVZ0, Q96C07, Q9UBQ8, Q9UIA2 | Gene names | CMPK, CMK, UCK, UMK, UMPK | |||
|
Domain Architecture |

|
|||||
| Description | UMP-CMP kinase (EC 2.7.4.14) (Cytidylate kinase) (Deoxycytidylate kinase) (Cytidine monophosphate kinase) (Uridine monophosphate/cytidine monophosphate kinase) (UMP/CMP kinase) (UMP/CMPK) (Uridine monophosphate kinase). | |||||
|
GEMI_MOUSE
|
||||||
| NC score | 0.075179 (rank : 24) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
GEMI_HUMAN
|
||||||
| NC score | 0.073828 (rank : 25) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75496, Q9H1Z1 | Gene names | GMNN | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.064400 (rank : 26) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
CDC37_HUMAN
|
||||||
| NC score | 0.063633 (rank : 27) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.059006 (rank : 28) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
NDUA5_MOUSE
|
||||||
| NC score | 0.059002 (rank : 29) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CPP6, Q9CY90, Q9D2P2, Q9D703, Q9D739 | Gene names | Ndufa5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 13 kDa-B subunit) (Complex I-13kD-B) (CI-13kD-B) (Complex I subunit B13). | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.058859 (rank : 30) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.057408 (rank : 31) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.057398 (rank : 32) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.055486 (rank : 33) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.053944 (rank : 34) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.052888 (rank : 35) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
STMN2_HUMAN
|
||||||
| NC score | 0.052508 (rank : 36) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q93045, O14952, Q6PK68 | Gene names | STMN2, SCG10, SCGN10 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
STMN2_MOUSE
|
||||||
| NC score | 0.052508 (rank : 37) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P55821 | Gene names | Stmn2, Scg10, Scgn10, Stmb2 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
NDK6_HUMAN
|
||||||
| NC score | 0.051715 (rank : 38) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75414, Q96E73, Q9BQ63 | Gene names | NME6 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoside diphosphate kinase 6 (EC 2.7.4.6) (NDK 6) (NDP kinase 6) (nm23-H6) (Inhibitor of p53-induced apoptosis-alpha) (IPIA-alpha). | |||||
|
NDK6_MOUSE
|
||||||
| NC score | 0.051708 (rank : 39) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88425, Q99M53 | Gene names | Nme6 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoside diphosphate kinase 6 (EC 2.7.4.6) (NDK 6) (NDP kinase 6) (nm23-M6). | |||||
|
STMN3_MOUSE
|
||||||
| NC score | 0.050390 (rank : 40) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O70166 | Gene names | Stmn3, Sclip | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-3 (SCG10-like protein) (SCG10-related protein HiAT3) (Hippocampus abundant transcript 3). | |||||
|
STMN3_HUMAN
|
||||||
| NC score | 0.050005 (rank : 41) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZ72, O75527, Q969Y4 | Gene names | STMN3, SCLIP | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-3 (SCG10-like protein). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.049374 (rank : 42) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
NDUA5_HUMAN
|
||||||
| NC score | 0.048980 (rank : 43) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16718, Q6IRX7 | Gene names | NDUFA5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 13 kDa-B subunit) (Complex I-13kD-B) (CI-13kD-B) (Complex I subunit B13). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.047647 (rank : 44) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.046415 (rank : 45) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.046236 (rank : 46) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
CT077_HUMAN
|
||||||
| NC score | 0.045521 (rank : 47) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQG5 | Gene names | C20orf77 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf77. | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.044925 (rank : 48) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.044543 (rank : 49) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.043446 (rank : 50) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.043119 (rank : 51) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.040763 (rank : 52) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.039580 (rank : 53) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.039457 (rank : 54) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
CCDC5_HUMAN
|
||||||
| NC score | 0.038949 (rank : 55) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96CS2, Q8N837 | Gene names | CCDC5, HEIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 5 (Enhancer of invasion-cluster) (HEI-C). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.038621 (rank : 56) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.036906 (rank : 57) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.036867 (rank : 58) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.036437 (rank : 59) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
CLH1_HUMAN
|
||||||
| NC score | 0.036381 (rank : 60) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00610, Q6N0A0, Q86TF2 | Gene names | CLTC, CLH17, KIAA0034 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 1 (CLH-17). | |||||
|
CLH_MOUSE
|
||||||
| NC score | 0.036376 (rank : 61) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68FD5 | Gene names | Cltc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin heavy chain. | |||||
|
THNSL_MOUSE
|
||||||
| NC score | 0.034356 (rank : 62) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.033297 (rank : 63) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.032171 (rank : 64) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.031388 (rank : 65) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.031113 (rank : 66) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.030803 (rank : 67) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
STX4_MOUSE
|
||||||
| NC score | 0.027914 (rank : 68) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70452 | Gene names | Stx4, Stx4a | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-4. | |||||
|
IF2M_MOUSE
|
||||||
| NC score | 0.027762 (rank : 69) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.027147 (rank : 70) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
ITPR2_MOUSE
|
||||||
| NC score | 0.026238 (rank : 71) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
ENKUR_HUMAN
|
||||||
| NC score | 0.026154 (rank : 72) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.025589 (rank : 73) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.025584 (rank : 74) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.025398 (rank : 75) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
RGNEF_MOUSE
|
||||||
| NC score | 0.024804 (rank : 76) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
ATAD1_HUMAN
|
||||||
| NC score | 0.024099 (rank : 77) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| NC score | 0.024099 (rank : 78) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.023230 (rank : 79) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
TRPM6_HUMAN
|
||||||
| NC score | 0.023089 (rank : 80) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.022179 (rank : 81) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.022178 (rank : 82) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
AR13B_HUMAN
|
||||||
| NC score | 0.020780 (rank : 83) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
ITPR2_HUMAN
|
||||||
| NC score | 0.020293 (rank : 84) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
LRC48_HUMAN
|
||||||
| NC score | 0.018753 (rank : 85) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H069, Q86SF9, Q86W73, Q8IWG0 | Gene names | LRRC48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 48. | |||||
|
KI13B_HUMAN
|
||||||
| NC score | 0.016983 (rank : 86) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.015214 (rank : 87) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
ALAT2_MOUSE
|
||||||
| NC score | 0.015165 (rank : 88) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGT5, Q8BVY7, Q8K1J3 | Gene names | Gpt2, Aat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alanine aminotransferase 2 (EC 2.6.1.2) (ALT2) (Glutamic--pyruvic transaminase 2) (GPT 2) (Glutamic--alanine transaminase 2). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.014602 (rank : 89) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.014273 (rank : 90) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.012092 (rank : 91) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.011285 (rank : 92) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
CT121_HUMAN
|
||||||
| NC score | 0.010772 (rank : 93) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTX7, Q5QPC1, Q9H1G2, Q9NQG8 | Gene names | C20orf121 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf121. | |||||
|
ABCG2_MOUSE
|
||||||
| NC score | 0.007758 (rank : 94) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TMS5, Q9R004, Q9Z1T0 | Gene names | Abcg2, Abcp, Bcrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family G member 2 (Breast cancer resistance protein 1 homolog). | |||||