Please be patient as the page loads
|
KCNK4_MOUSE
|
||||||
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCNK4_MOUSE
|
||||||
| θ value | 8.84838e-173 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | 3.28708e-135 (rank : 2) | NC score | 0.984754 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | 3.12697e-77 (rank : 3) | NC score | 0.963420 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | 5.8972e-76 (rank : 4) | NC score | 0.963271 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | 2.4834e-66 (rank : 5) | NC score | 0.953985 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 4.13158e-37 (rank : 6) | NC score | 0.926790 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | 3.86705e-35 (rank : 7) | NC score | 0.942563 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 8) | NC score | 0.919199 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 9) | NC score | 0.896030 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 10) | NC score | 0.848998 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK1_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 11) | NC score | 0.874501 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 12) | NC score | 0.849493 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK9_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 13) | NC score | 0.853804 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 14) | NC score | 0.858395 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNK1_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 15) | NC score | 0.872193 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 16) | NC score | 0.853658 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNKD_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 17) | NC score | 0.790999 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKD_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 18) | NC score | 0.783818 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKC_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 19) | NC score | 0.795262 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 20) | NC score | 0.819799 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNK7_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 21) | NC score | 0.803023 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 22) | NC score | 0.203466 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 23) | NC score | 0.203763 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 24) | NC score | 0.193867 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 25) | NC score | 0.193814 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.034196 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.037095 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.008440 (rank : 130) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.185188 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.185399 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.017677 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.148131 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.160256 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.147490 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNT1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.193549 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KCNT1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.183320 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZPR4, Q8C3E7 | Gene names | Kcnt1, Kiaa1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1. | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.158648 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.156996 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.119950 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.118875 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.128531 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.152788 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.152683 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.127291 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.127314 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.126436 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.126484 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.159051 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.123754 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.123263 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.087966 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.008395 (rank : 131) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.023724 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.099309 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.152361 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.154005 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.074211 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
NRAM1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.048644 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41251 | Gene names | Slc11a1, Bcg, Ity, Lsh, Nramp1 | |||
|
Domain Architecture |
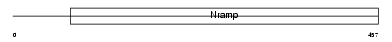
|
|||||
| Description | Natural resistance-associated macrophage protein 1 (NRAMP 1). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.017882 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.124644 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.016059 (rank : 119) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.020357 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
GLIS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | -0.000430 (rank : 145) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
KCNN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.082353 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.084871 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.077435 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.076817 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.022935 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.027446 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
EPHAA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | -0.000619 (rank : 146) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5JZY3, Q6NW42 | Gene names | EPHA10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephrin type-A receptor 10 precursor (EC 2.7.10.1). | |||||
|
NRAM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.049365 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49279 | Gene names | SLC11A1, NRAMP, NRAMP1 | |||
|
Domain Architecture |
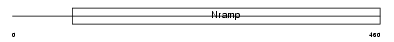
|
|||||
| Description | Natural resistance-associated macrophage protein 1 (NRAMP 1). | |||||
|
PPIP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.012109 (rank : 123) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H939 | Gene names | PSTPIP2 | |||
|
Domain Architecture |
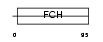
|
|||||
| Description | Proline-serine-threonine phosphatase-interacting protein 2. | |||||
|
APBA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.009590 (rank : 128) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.096716 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.127181 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.088106 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.088044 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
NRAM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.041618 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49281, O43288, O60932, Q96J35 | Gene names | SLC11A2, NRAMP2 | |||
|
Domain Architecture |
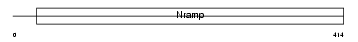
|
|||||
| Description | Natural resistance-associated macrophage protein 2 (NRAMP 2) (Divalent metal transporter 1) (DMT1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.015916 (rank : 120) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
IF4B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.029160 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.025005 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.084294 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
NRAM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.040325 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49282, O54903 | Gene names | Slc11a2, Nramp2 | |||
|
Domain Architecture |
|
|||||
| Description | Natural resistance-associated macrophage protein 2 (NRAMP 2). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.018952 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SRC8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.006784 (rank : 136) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
|
|||||
| Description | Src substrate cortactin. | |||||
|
STK6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | -0.002312 (rank : 147) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97477, O35624, Q91YU4 | Gene names | Stk6, Ark1, Aurka, Ayk1, Iak1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Aurora-family kinase 1) (Aurora/IPL1-related kinase 1) (Ipl1- and aurora-related kinase 1) (Aurora-A) (Serine/threonine-protein kinase Ayk1). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.010415 (rank : 125) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
BAP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.017466 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92560, Q6LEM0, Q7Z5E8 | Gene names | BAP1, KIAA0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Cerebral protein 6). | |||||
|
BAP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.017491 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99PU7, Q6ZQE6 | Gene names | Bap1, Kiaa0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Ubiquitin C-terminal hydrolase X4) (UCH-X4). | |||||
|
INP5E_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.011278 (rank : 124) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.096493 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.082038 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.117603 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.117001 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.094852 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.005967 (rank : 139) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.006987 (rank : 134) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
YIF1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.020462 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91XB7, Q9CWB2 | Gene names | Yif1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIF1A (YIP1-interacting factor homolog A). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.014617 (rank : 121) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CIKS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.017230 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43734, Q5R3A3, Q9H5W2, Q9H6Y3, Q9NS14, Q9UG72 | Gene names | TRAF3IP2, C6orf4, C6orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adapter protein CIKS (Connection to IKK and SAPK/JNK) (TRAF3- interacting protein 2) (Nuclear factor NF-kappa-B activator 1) (ACT1). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.009728 (rank : 127) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
K1849_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.007746 (rank : 132) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.086176 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.086100 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
L2GL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.006566 (rank : 137) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15334, O00188, Q58F11, Q86UK6 | Gene names | LLGL1, HUGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(2) giant larvae protein homolog 1 (LLGL) (Human homolog to the D-lgl gene) (Hugl-1) (DLG4). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.009959 (rank : 126) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | -0.000241 (rank : 144) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.006057 (rank : 138) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
YIF1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.031712 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95070, Q96G83, Q9BVD0 | Gene names | YIF1A, 54TM, HYIF1P, YIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIF1A (YIP1-interacting factor homolog A) (54TMp). | |||||
|
AQP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.005630 (rank : 140) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94778, Q8IUU3, Q9UIA4 | Gene names | AQP8 | |||
|
Domain Architecture |
|
|||||
| Description | Aquaporin-8 (AQP-8). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.012153 (rank : 122) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.019715 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CSPG5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.009022 (rank : 129) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
HIRA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.002670 (rank : 142) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61666, O08845, Q62365 | Gene names | Hira, Tuple1 | |||
|
Domain Architecture |
|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.007718 (rank : 133) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.000752 (rank : 143) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.006952 (rank : 135) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.002796 (rank : 141) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051720 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.050891 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.050601 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.050211 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.084624 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.084667 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.090049 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.089981 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.091411 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.093305 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.104106 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.093386 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.097901 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.095174 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.086081 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.086134 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.086639 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.079120 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.086669 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.086816 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.083384 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.064371 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.064410 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.071720 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.076468 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.076473 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.115370 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCNN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.116199 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.067178 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 8.84838e-173 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.984754 (rank : 2) | θ value | 3.28708e-135 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.963420 (rank : 3) | θ value | 3.12697e-77 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.963271 (rank : 4) | θ value | 5.8972e-76 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.953985 (rank : 5) | θ value | 2.4834e-66 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.942563 (rank : 6) | θ value | 3.86705e-35 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.926790 (rank : 7) | θ value | 4.13158e-37 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.919199 (rank : 8) | θ value | 1.28732e-30 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.896030 (rank : 9) | θ value | 4.89182e-30 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK1_HUMAN
|
||||||
| NC score | 0.874501 (rank : 10) | θ value | 2.35151e-24 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNK1_MOUSE
|
||||||
| NC score | 0.872193 (rank : 11) | θ value | 1.16704e-23 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.858395 (rank : 12) | θ value | 8.93572e-24 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNK9_HUMAN
|
||||||
| NC score | 0.853804 (rank : 13) | θ value | 8.93572e-24 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.853658 (rank : 14) | θ value | 2.69047e-20 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.849493 (rank : 15) | θ value | 5.23862e-24 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.848998 (rank : 16) | θ value | 3.62785e-25 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.819799 (rank : 17) | θ value | 1.2105e-12 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNK7_HUMAN
|
||||||
| NC score | 0.803023 (rank : 18) | θ value | 1.80886e-08 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCNKC_HUMAN
|
||||||
| NC score | 0.795262 (rank : 19) | θ value | 3.76295e-14 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
KCNKD_MOUSE
|
||||||
| NC score | 0.790999 (rank : 20) | θ value | 6.85773e-16 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKD_HUMAN
|
||||||
| NC score | 0.783818 (rank : 21) | θ value | 2.60593e-15 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.203763 (rank : 22) | θ value | 0.00298849 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.203466 (rank : 23) | θ value | 0.00298849 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.193867 (rank : 24) | θ value | 0.0252991 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.193814 (rank : 25) | θ value | 0.0252991 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNT1_HUMAN
|
||||||
| NC score | 0.193549 (rank : 26) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.185399 (rank : 27) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.185188 (rank : 28) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNT1_MOUSE
|
||||||
| NC score | 0.183320 (rank : 29) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZPR4, Q8C3E7 | Gene names | Kcnt1, Kiaa1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1. | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.160256 (rank : 30) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.159051 (rank : 31) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.158648 (rank : 32) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.156996 (rank : 33) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.154005 (rank : 34) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.152788 (rank : 35) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.152683 (rank : 36) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.152361 (rank : 37) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.148131 (rank : 38) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.147490 (rank : 39) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.128531 (rank : 40) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.127314 (rank : 41) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.127291 (rank : 42) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.127181 (rank : 43) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.126484 (rank : 44) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.126436 (rank : 45) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.124644 (rank : 46) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.123754 (rank : 47) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.123263 (rank : 48) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.119950 (rank : 49) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.118875 (rank : 50) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.117603 (rank : 51) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.117001 (rank : 52) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNN4_MOUSE
|
||||||
| NC score | 0.116199 (rank : 53) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNN4_HUMAN
|
||||||
| NC score | 0.115370 (rank : 54) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.104106 (rank : 55) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.099309 (rank : 56) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.097901 (rank : 57) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.096716 (rank : 58) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.096493 (rank : 59) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.095174 (rank : 60) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.094852 (rank : 61) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.093386 (rank : 62) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.093305 (rank : 63) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.091411 (rank : 64) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.090049 (rank : 65) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.089981 (rank : 66) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.088106 (rank : 67) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.088044 (rank : 68) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.087966 (rank : 69) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.086816 (rank : 70) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.086669 (rank : 71) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.086639 (rank : 72) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.086176 (rank : 73) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.086134 (rank : 74) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.086100 (rank : 75) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.086081 (rank : 76) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNN1_MOUSE
|
||||||
| NC score | 0.084871 (rank : 77) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.084667 (rank : 78) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.084624 (rank : 79) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.084294 (rank : 80) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.083384 (rank : 81) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNN1_HUMAN
|
||||||
| NC score | 0.082353 (rank : 82) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.082038 (rank : 83) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.079120 (rank : 84) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.077435 (rank : 85) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.076817 (rank : 86) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.076473 (rank : 87) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_HUMAN
|
||||||
| NC score | 0.076468 (rank : 88) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.074211 (rank : 89) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.071720 (rank : 90) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.067178 (rank : 91) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.064410 (rank : 92) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.064371 (rank : 93) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.051720 (rank : 94) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA3_MOUSE
|
||||||
| NC score | 0.050891 (rank : 95) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.050601 (rank : 96) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.050211 (rank : 97) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
NRAM1_HUMAN
|
||||||
| NC score | 0.049365 (rank : 98) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49279 | Gene names | SLC11A1, NRAMP, NRAMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Natural resistance-associated macrophage protein 1 (NRAMP 1). | |||||
|
NRAM1_MOUSE
|
||||||
| NC score | 0.048644 (rank : 99) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41251 | Gene names | Slc11a1, Bcg, Ity, Lsh, Nramp1 | |||
|
Domain Architecture |

|
|||||
| Description | Natural resistance-associated macrophage protein 1 (NRAMP 1). | |||||
|
NRAM2_HUMAN
|
||||||
| NC score | 0.041618 (rank : 100) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49281, O43288, O60932, Q96J35 | Gene names | SLC11A2, NRAMP2 | |||
|
Domain Architecture |

|
|||||
| Description | Natural resistance-associated macrophage protein 2 (NRAMP 2) (Divalent metal transporter 1) (DMT1). | |||||
|
NRAM2_MOUSE
|
||||||
| NC score | 0.040325 (rank : 101) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49282, O54903 | Gene names | Slc11a2, Nramp2 | |||
|
Domain Architecture |

|
|||||
| Description | Natural resistance-associated macrophage protein 2 (NRAMP 2). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.037095 (rank : 102) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.034196 (rank : 103) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
YIF1A_HUMAN
|
||||||
| NC score | 0.031712 (rank : 104) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95070, Q96G83, Q9BVD0 | Gene names | YIF1A, 54TM, HYIF1P, YIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIF1A (YIP1-interacting factor homolog A) (54TMp). | |||||
|
IF4B_HUMAN
|
||||||
| NC score | 0.029160 (rank : 105) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.027446 (rank : 106) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.025005 (rank : 107) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.023724 (rank : 108) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.022935 (rank : 109) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
YIF1A_MOUSE
|
||||||
| NC score | 0.020462 (rank : 110) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91XB7, Q9CWB2 | Gene names | Yif1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIF1A (YIP1-interacting factor homolog A). | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.020357 (rank : 111) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.019715 (rank : 112) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.018952 (rank : 113) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.017882 (rank : 114) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.017677 (rank : 115) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
BAP1_MOUSE
|
||||||
| NC score | 0.017491 (rank : 116) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99PU7, Q6ZQE6 | Gene names | Bap1, Kiaa0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Ubiquitin C-terminal hydrolase X4) (UCH-X4). | |||||
|
BAP1_HUMAN
|
||||||
| NC score | 0.017466 (rank : 117) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92560, Q6LEM0, Q7Z5E8 | Gene names | BAP1, KIAA0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Cerebral protein 6). | |||||
|
CIKS_HUMAN
|
||||||
| NC score | 0.017230 (rank : 118) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43734, Q5R3A3, Q9H5W2, Q9H6Y3, Q9NS14, Q9UG72 | Gene names | TRAF3IP2, C6orf4, C6orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adapter protein CIKS (Connection to IKK and SAPK/JNK) (TRAF3- interacting protein 2) (Nuclear factor NF-kappa-B activator 1) (ACT1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.016059 (rank : 119) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.015916 (rank : 120) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.014617 (rank : 121) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.012153 (rank : 122) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
PPIP2_HUMAN
|
||||||
| NC score | 0.012109 (rank : 123) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H939 | Gene names | PSTPIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Proline-serine-threonine phosphatase-interacting protein 2. | |||||
|
INP5E_HUMAN
|
||||||
| NC score | 0.011278 (rank : 124) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.010415 (rank : 125) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.009959 (rank : 126) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.009728 (rank : 127) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.009590 (rank : 128) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
CSPG5_MOUSE
|
||||||
| NC score | 0.009022 (rank : 129) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.008440 (rank : 130) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.008395 (rank : 131) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
K1849_MOUSE
|
||||||
| NC score | 0.007746 (rank : 132) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.007718 (rank : 133) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.006987 (rank : 134) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.006952 (rank : 135) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SRC8_MOUSE
|
||||||
| NC score | 0.006784 (rank : 136) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
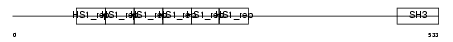
|
|||||
| Description | Src substrate cortactin. | |||||
|
L2GL1_HUMAN
|
||||||
| NC score | 0.006566 (rank : 137) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15334, O00188, Q58F11, Q86UK6 | Gene names | LLGL1, HUGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(2) giant larvae protein homolog 1 (LLGL) (Human homolog to the D-lgl gene) (Hugl-1) (DLG4). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.006057 (rank : 138) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.005967 (rank : 139) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
AQP8_HUMAN
|
||||||
| NC score | 0.005630 (rank : 140) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94778, Q8IUU3, Q9UIA4 | Gene names | AQP8 | |||
|
Domain Architecture |

|
|||||
| Description | Aquaporin-8 (AQP-8). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.002796 (rank : 141) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
HIRA_MOUSE
|
||||||
| NC score | 0.002670 (rank : 142) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61666, O08845, Q62365 | Gene names | Hira, Tuple1 | |||
|
Domain Architecture |

|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.000752 (rank : 143) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | -0.000241 (rank : 144) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
GLIS3_MOUSE
|
||||||
| NC score | -0.000430 (rank : 145) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
EPHAA_HUMAN
|
||||||
| NC score | -0.000619 (rank : 146) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5JZY3, Q6NW42 | Gene names | EPHA10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephrin type-A receptor 10 precursor (EC 2.7.10.1). | |||||
|
STK6_MOUSE
|
||||||
| NC score | -0.002312 (rank : 147) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97477, O35624, Q91YU4 | Gene names | Stk6, Ark1, Aurka, Ayk1, Iak1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Aurora-family kinase 1) (Aurora/IPL1-related kinase 1) (Ipl1- and aurora-related kinase 1) (Aurora-A) (Serine/threonine-protein kinase Ayk1). | |||||