Please be patient as the page loads
|
BAP1_MOUSE
|
||||||
| SwissProt Accessions | Q99PU7, Q6ZQE6 | Gene names | Bap1, Kiaa0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Ubiquitin C-terminal hydrolase X4) (UCH-X4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BAP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985754 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q92560, Q6LEM0, Q7Z5E8 | Gene names | BAP1, KIAA0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Cerebral protein 6). | |||||
|
BAP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q99PU7, Q6ZQE6 | Gene names | Bap1, Kiaa0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Ubiquitin C-terminal hydrolase X4) (UCH-X4). | |||||
|
UCHL5_HUMAN
|
||||||
| θ value | 7.51208e-47 (rank : 3) | NC score | 0.832833 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y5K5, Q5LJA6, Q5LJA7, Q8TBS4, Q96BJ9, Q9H1W5, Q9P0I3, Q9UQN2 | Gene names | UCHL5, UCH37 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH- L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37). | |||||
|
UCHL5_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 4) | NC score | 0.833560 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUP7, Q9CVJ4, Q9CXZ3, Q9R107 | Gene names | Uchl5, Uch37 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH- L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37). | |||||
|
UCHL3_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 5) | NC score | 0.554338 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15374, Q5TBK8, Q6IBE9 | Gene names | UCHL3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L3 (EC 3.4.19.12) (UCH- L3) (Ubiquitin thioesterase L3). | |||||
|
UCHL3_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 6) | NC score | 0.555008 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JKB1, Q9EQX7 | Gene names | Uchl3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L3 (EC 3.4.19.12) (UCH- L3) (Ubiquitin thioesterase L3). | |||||
|
UCHL4_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 7) | NC score | 0.548704 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58321 | Gene names | Uchl4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L4 (EC 3.4.19.12) (UCH- L4) (Ubiquitin thioesterase L4). | |||||
|
UCHL1_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 8) | NC score | 0.527558 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0P9, Q9R122 | Gene names | Uchl1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L1 (EC 3.4.19.12) (EC 6.-.-.-) (UCH-L1) (Ubiquitin thioesterase L1) (Neuron cytoplasmic protein 9.5) (PGP 9.5) (PGP9.5). | |||||
|
UCHL1_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 9) | NC score | 0.513085 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P09936, Q71UM0 | Gene names | UCHL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L1 (EC 3.4.19.12) (EC 6.-.-.-) (UCH-L1) (Ubiquitin thioesterase L1) (Neuron cytoplasmic protein 9.5) (PGP 9.5) (PGP9.5). | |||||
|
K1958_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 10) | NC score | 0.114955 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C4P0, Q8BN62, Q8K1X8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958 homolog. | |||||
|
E2F3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.046953 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |
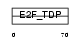
|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
K1958_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.098242 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.040733 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.049473 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TEX2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.090299 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IWB9, Q6AHZ5, Q8N3L0, Q9C0C5 | Gene names | TEX2, KIAA1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
BRSK2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.011815 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.070354 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
FURIN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.031420 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09958, Q14336, Q6LBS3 | Gene names | FURIN, FUR, PACE | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.072692 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.035251 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.056776 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.067662 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.063409 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.014578 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.043762 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TEX2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.080744 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
VCIP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.069217 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JH7, Q504T4, Q86T93, Q86W01, Q8N3A9, Q9H5R8 | Gene names | VCPIP1, KIAA1850, VCIP135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.060174 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.043984 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.048127 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
VCIP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.067100 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CDG3, Q7TMU9, Q8BP90 | Gene names | Vcpip1, Vcip135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
BFSP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.025667 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 508 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12934, O43595, O76034, Q9HBX4 | Gene names | BFSP1 | |||
|
Domain Architecture |
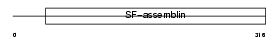
|
|||||
| Description | Filensin (Beaded filament structural protein 1) (Lens fiber cell beaded-filament structural protein CP 115) (CP115) (Lens intermediate filament-like heavy) (LIFL-H). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.026618 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.013639 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.054285 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.023159 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.055430 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.055024 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
DAG1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.042133 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62165, Q61094, Q61141, Q61497 | Gene names | Dag1, Dag-1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
DESP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.011665 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
GATA6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.027709 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.037718 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.027592 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
ADRM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.035534 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |
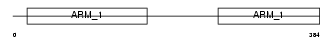
|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
CCD94_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.039027 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D6J3 | Gene names | Ccdc94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 94. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.049551 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.032988 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.023995 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.059061 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.046683 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.036988 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SALL4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.007691 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJQ4 | Gene names | SALL4 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 4 (Zinc finger protein SALL4). | |||||
|
SON_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.031514 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.032007 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.019259 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.004124 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CI093_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.015578 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CT055_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.029849 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BQ89 | Gene names | C20orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf55. | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.020403 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.009810 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.012773 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.009658 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.039163 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.031642 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.010204 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.019074 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
DACH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.016945 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
GPR64_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.013093 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
HNF3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.011405 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35582, Q61108 | Gene names | Foxa1, Hnf3a, Tcf-3a, Tcf3a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
KCNK4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.017491 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.007243 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.014687 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MAVS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.035210 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.031882 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.029443 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.005981 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.035509 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.046869 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.009455 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ADA19_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.005889 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35674 | Gene names | Adam19, Mltnb | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta). | |||||
|
AIM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.017959 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.009098 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
APC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.026436 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.010888 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.004036 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.019776 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
INADL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.006202 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.018998 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KCC2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.006769 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13554, O95437, O95438, O95599, Q9UGH7, Q9UGH8, Q9UGH9, Q9UNX0, Q9UNX7, Q9UP00, Q9Y5N4, Q9Y6F4 | Gene names | CAMK2B, CAM2, CAMKB | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II beta chain (EC 2.7.11.17) (CaM-kinase II beta chain) (CaM kinase II subunit beta) (CaMK-II subunit beta). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.021926 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.018005 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.008246 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.013960 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SALL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.005677 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
SALL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.006379 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ER74, Q920R5 | Gene names | Sall1, Sal3 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein Spalt-3) (Sal-3) (MSal-3). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.015602 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
TRIB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.008465 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RU8, O15180, Q9H2Y8 | Gene names | TRIB1, C8FW, GIG2, TRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tribbles homolog 1 (TRB-1) (SKIP1) (G-protein-coupled receptor-induced protein 2) (GIG-2). | |||||
|
TSH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.015102 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
XRCC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.018343 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924T3, Q8BKC9, Q8BU02, Q9D6U6 | Gene names | Xrcc4 | |||
|
Domain Architecture |
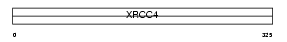
|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.034122 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.025402 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.016180 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
CD5R2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.012472 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13319, Q4ZFW6 | Gene names | CDK5R2, NCK5AI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
DACT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.022555 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
EGR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.003791 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P18146 | Gene names | EGR1, ZNF225 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A) (Transcription factor ETR103) (Zinc finger protein 225) (AT225). | |||||
|
ELOA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.011472 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
GLE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.017193 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
HNF3G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.005249 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35584 | Gene names | Foxa3, Hnf3g, Tcf-3g, Tcf3g | |||
|
Domain Architecture |
|
|||||
| Description | Hepatocyte nuclear factor 3-gamma (HNF-3G) (Forkhead box protein A3). | |||||
|
IL2RA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.014780 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01589 | Gene names | IL2RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor alpha chain precursor (IL-2 receptor alpha subunit) (IL-2-RA) (IL2-RA) (p55) (TAC antigen) (CD25 antigen). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.012743 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.012471 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.021444 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.011408 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.017198 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.015785 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TRIB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.008101 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4K4, Q8BFS7, Q8BJR9, Q8BZX3, Q91W04 | Gene names | Trib1, Trb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tribbles homolog 1 (TRB-1). | |||||
|
TSKS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.019400 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54887 | Gene names | Stk22s1, Tsks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
ZBT40_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | -0.000482 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
BAP1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q99PU7, Q6ZQE6 | Gene names | Bap1, Kiaa0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Ubiquitin C-terminal hydrolase X4) (UCH-X4). | |||||
|
BAP1_HUMAN
|
||||||
| NC score | 0.985754 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q92560, Q6LEM0, Q7Z5E8 | Gene names | BAP1, KIAA0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Cerebral protein 6). | |||||
|
UCHL5_MOUSE
|
||||||
| NC score | 0.833560 (rank : 3) | θ value | 1.28137e-46 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUP7, Q9CVJ4, Q9CXZ3, Q9R107 | Gene names | Uchl5, Uch37 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH- L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37). | |||||
|
UCHL5_HUMAN
|
||||||
| NC score | 0.832833 (rank : 4) | θ value | 7.51208e-47 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y5K5, Q5LJA6, Q5LJA7, Q8TBS4, Q96BJ9, Q9H1W5, Q9P0I3, Q9UQN2 | Gene names | UCHL5, UCH37 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH- L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37). | |||||
|
UCHL3_MOUSE
|
||||||
| NC score | 0.555008 (rank : 5) | θ value | 1.33837e-11 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JKB1, Q9EQX7 | Gene names | Uchl3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L3 (EC 3.4.19.12) (UCH- L3) (Ubiquitin thioesterase L3). | |||||
|
UCHL3_HUMAN
|
||||||
| NC score | 0.554338 (rank : 6) | θ value | 1.33837e-11 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15374, Q5TBK8, Q6IBE9 | Gene names | UCHL3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L3 (EC 3.4.19.12) (UCH- L3) (Ubiquitin thioesterase L3). | |||||
|
UCHL4_MOUSE
|
||||||
| NC score | 0.548704 (rank : 7) | θ value | 2.28291e-11 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58321 | Gene names | Uchl4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L4 (EC 3.4.19.12) (UCH- L4) (Ubiquitin thioesterase L4). | |||||
|
UCHL1_MOUSE
|
||||||
| NC score | 0.527558 (rank : 8) | θ value | 2.13673e-09 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0P9, Q9R122 | Gene names | Uchl1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L1 (EC 3.4.19.12) (EC 6.-.-.-) (UCH-L1) (Ubiquitin thioesterase L1) (Neuron cytoplasmic protein 9.5) (PGP 9.5) (PGP9.5). | |||||
|
UCHL1_HUMAN
|
||||||
| NC score | 0.513085 (rank : 9) | θ value | 5.26297e-08 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P09936, Q71UM0 | Gene names | UCHL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L1 (EC 3.4.19.12) (EC 6.-.-.-) (UCH-L1) (Ubiquitin thioesterase L1) (Neuron cytoplasmic protein 9.5) (PGP 9.5) (PGP9.5). | |||||
|
K1958_MOUSE
|
||||||
| NC score | 0.114955 (rank : 10) | θ value | 0.00102713 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C4P0, Q8BN62, Q8K1X8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958 homolog. | |||||
|
K1958_HUMAN
|
||||||
| NC score | 0.098242 (rank : 11) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
TEX2_HUMAN
|
||||||
| NC score | 0.090299 (rank : 12) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IWB9, Q6AHZ5, Q8N3L0, Q9C0C5 | Gene names | TEX2, KIAA1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
TEX2_MOUSE
|
||||||
| NC score | 0.080744 (rank : 13) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.072692 (rank : 14) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.070354 (rank : 15) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
VCIP1_HUMAN
|
||||||
| NC score | 0.069217 (rank : 16) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JH7, Q504T4, Q86T93, Q86W01, Q8N3A9, Q9H5R8 | Gene names | VCPIP1, KIAA1850, VCIP135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.067662 (rank : 17) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
VCIP1_MOUSE
|
||||||
| NC score | 0.067100 (rank : 18) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CDG3, Q7TMU9, Q8BP90 | Gene names | Vcpip1, Vcip135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.063409 (rank : 19) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.060174 (rank : 20) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.059061 (rank : 21) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.056776 (rank : 22) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.055430 (rank : 23) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.055024 (rank : 24) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.054285 (rank : 25) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.049551 (rank : 26) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.049473 (rank : 27) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.048127 (rank : 28) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
E2F3_HUMAN
|
||||||
| NC score | 0.046953 (rank : 29) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.046869 (rank : 30) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.046683 (rank : 31) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.043984 (rank : 32) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.043762 (rank : 33) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
DAG1_MOUSE
|
||||||
| NC score | 0.042133 (rank : 34) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62165, Q61094, Q61141, Q61497 | Gene names | Dag1, Dag-1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.040733 (rank : 35) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.039163 (rank : 36) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CCD94_MOUSE
|
||||||
| NC score | 0.039027 (rank : 37) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D6J3 | Gene names | Ccdc94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 94. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.037718 (rank : 38) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.036988 (rank : 39) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
ADRM1_HUMAN
|
||||||
| NC score | 0.035534 (rank : 40) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.035509 (rank : 41) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.035251 (rank : 42) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAVS_MOUSE
|
||||||
| NC score | 0.035210 (rank : 43) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.034122 (rank : 44) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.032988 (rank : 45) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.032007 (rank : 46) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.031882 (rank : 47) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.031642 (rank : 48) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.031514 (rank : 49) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
FURIN_HUMAN
|
||||||
| NC score | 0.031420 (rank : 50) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09958, Q14336, Q6LBS3 | Gene names | FURIN, FUR, PACE | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme). | |||||
|
CT055_HUMAN
|
||||||
| NC score | 0.029849 (rank : 51) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BQ89 | Gene names | C20orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf55. | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.029443 (rank : 52) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
GATA6_HUMAN
|
||||||
| NC score | 0.027709 (rank : 53) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.027592 (rank : 54) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.026618 (rank : 55) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.026436 (rank : 56) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
BFSP1_HUMAN
|
||||||
| NC score | 0.025667 (rank : 57) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 508 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12934, O43595, O76034, Q9HBX4 | Gene names | BFSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Filensin (Beaded filament structural protein 1) (Lens fiber cell beaded-filament structural protein CP 115) (CP115) (Lens intermediate filament-like heavy) (LIFL-H). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.025402 (rank : 58) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.023995 (rank : 59) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.023159 (rank : 60) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
DACT1_HUMAN
|
||||||
| NC score | 0.022555 (rank : 61) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.021926 (rank : 62) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.021444 (rank : 63) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.020403 (rank : 64) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.019776 (rank : 65) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
TSKS_MOUSE
|
||||||
| NC score | 0.019400 (rank : 66) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54887 | Gene names | Stk22s1, Tsks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.019259 (rank : 67) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.019074 (rank : 68) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.018998 (rank : 69) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
XRCC4_MOUSE
|
||||||
| NC score | 0.018343 (rank : 70) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924T3, Q8BKC9, Q8BU02, Q9D6U6 | Gene names | Xrcc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.018005 (rank : 71) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
AIM1_HUMAN
|
||||||
| NC score | 0.017959 (rank : 72) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.017491 (rank : 73) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.017198 (rank : 74) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
GLE1_MOUSE
|
||||||
| NC score | 0.017193 (rank : 75) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
DACH1_HUMAN
|
||||||
| NC score | 0.016945 (rank : 76) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.016180 (rank : 77) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.015785 (rank : 78) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.015602 (rank : 79) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.015578 (rank : 80) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
TSH1_HUMAN
|
||||||
| NC score | 0.015102 (rank : 81) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
IL2RA_HUMAN
|
||||||
| NC score | 0.014780 (rank : 82) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01589 | Gene names | IL2RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor alpha chain precursor (IL-2 receptor alpha subunit) (IL-2-RA) (IL2-RA) (p55) (TAC antigen) (CD25 antigen). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.014687 (rank : 83) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.014578 (rank : 84) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.013960 (rank : 85) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.013639 (rank : 86) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
GPR64_HUMAN
|
||||||
| NC score | 0.013093 (rank : 87) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.012773 (rank : 88) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.012743 (rank : 89) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
CD5R2_HUMAN
|
||||||
| NC score | 0.012472 (rank : 90) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13319, Q4ZFW6 | Gene names | CDK5R2, NCK5AI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
NCA11_MOUSE
|
||||||
| NC score | 0.012471 (rank : 91) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
BRSK2_HUMAN
|
||||||
| NC score | 0.011815 (rank : 92) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.011665 (rank : 93) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
ELOA1_MOUSE
|
||||||
| NC score | 0.011472 (rank : 94) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.011408 (rank : 95) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
HNF3A_MOUSE
|
||||||
| NC score | 0.011405 (rank : 96) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35582, Q61108 | Gene names | Foxa1, Hnf3a, Tcf-3a, Tcf3a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.010888 (rank : 97) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.010204 (rank : 98) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.009810 (rank : 99) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | 0.009658 (rank : 100) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.009455 (rank : 101) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.009098 (rank : 102) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
TRIB1_HUMAN
|
||||||
| NC score | 0.008465 (rank : 103) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RU8, O15180, Q9H2Y8 | Gene names | TRIB1, C8FW, GIG2, TRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tribbles homolog 1 (TRB-1) (SKIP1) (G-protein-coupled receptor-induced protein 2) (GIG-2). | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.008246 (rank : 104) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
TRIB1_MOUSE
|
||||||
| NC score | 0.008101 (rank : 105) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4K4, Q8BFS7, Q8BJR9, Q8BZX3, Q91W04 | Gene names | Trib1, Trb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tribbles homolog 1 (TRB-1). | |||||
|
SALL4_HUMAN
|
||||||
| NC score | 0.007691 (rank : 106) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJQ4 | Gene names | SALL4 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 4 (Zinc finger protein SALL4). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.007243 (rank : 107) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
KCC2B_HUMAN
|
||||||
| NC score | 0.006769 (rank : 108) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13554, O95437, O95438, O95599, Q9UGH7, Q9UGH8, Q9UGH9, Q9UNX0, Q9UNX7, Q9UP00, Q9Y5N4, Q9Y6F4 | Gene names | CAMK2B, CAM2, CAMKB | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II beta chain (EC 2.7.11.17) (CaM-kinase II beta chain) (CaM kinase II subunit beta) (CaMK-II subunit beta). | |||||
|
SALL1_MOUSE
|
||||||
| NC score | 0.006379 (rank : 109) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ER74, Q920R5 | Gene names | Sall1, Sal3 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein Spalt-3) (Sal-3) (MSal-3). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.006202 (rank : 110) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.005981 (rank : 111) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
ADA19_MOUSE
|
||||||
| NC score | 0.005889 (rank : 112) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35674 | Gene names | Adam19, Mltnb | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta). | |||||
|
SALL1_HUMAN
|
||||||
| NC score | 0.005677 (rank : 113) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
HNF3G_MOUSE
|
||||||
| NC score | 0.005249 (rank : 114) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35584 | Gene names | Foxa3, Hnf3g, Tcf-3g, Tcf3g | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-gamma (HNF-3G) (Forkhead box protein A3). | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.004124 (rank : 115) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
EGR1_MOUSE
|
||||||
| NC score | 0.004036 (rank : 116) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
EGR1_HUMAN
|
||||||
| NC score | 0.003791 (rank : 117) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P18146 | Gene names | EGR1, ZNF225 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A) (Transcription factor ETR103) (Zinc finger protein 225) (AT225). | |||||
|
ZBT40_HUMAN
|
||||||
| NC score | -0.000482 (rank : 118) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||