Please be patient as the page loads
|
CD5R2_HUMAN
|
||||||
| SwissProt Accessions | Q13319, Q4ZFW6 | Gene names | CDK5R2, NCK5AI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CD5R2_MOUSE
|
||||||
| θ value | 2.51683e-135 (rank : 1) | NC score | 0.985870 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35926, O35277 | Gene names | Cdk5r2, Nck5ai | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
CD5R2_HUMAN
|
||||||
| θ value | 3.5201e-129 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13319, Q4ZFW6 | Gene names | CDK5R2, NCK5AI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
CD5R1_MOUSE
|
||||||
| θ value | 1.36269e-64 (rank : 3) | NC score | 0.935963 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61809, Q62938 | Gene names | Cdk5r1, Cdk5r, Nck5a | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
CD5R1_HUMAN
|
||||||
| θ value | 2.17557e-62 (rank : 4) | NC score | 0.934390 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15078 | Gene names | CDK5R1, CDK5R, NCK5A | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 5) | NC score | 0.070373 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.102618 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.072039 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.082878 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.090563 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.061102 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.061301 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.061248 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.014371 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.020034 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.017525 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
CN037_MOUSE
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.044983 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.043898 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.045947 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.046086 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.052797 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.070787 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.058615 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
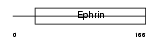
EFNB3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.032459 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35393 | Gene names | Efnb3 | |||
|
Domain Architecture |
|
|||||
| Description | Ephrin-B3 precursor. | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.046373 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.042645 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
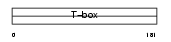
TBX18_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.012599 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
PUNC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.007745 (rank : 57) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BQC3, O70246, Q792T2, Q9Z2S6 | Gene names | Punc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
SCAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.029965 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
SON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.039585 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ADA19_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.008936 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.022102 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.037346 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.035196 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.016189 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
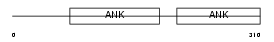
PP16A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.011358 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
SON_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.049239 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
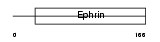
EFNB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.027820 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15768, O00680, Q8TBH7, Q92875 | Gene names | EFNB3, EPLG8, LERK8 | |||
|
Domain Architecture |
|
|||||
| Description | Ephrin-B3 precursor (EPH-related receptor tyrosine kinase ligand 8) (LERK-8) (EPH-related receptor transmembrane ligand ELK-L3). | |||||
|
PAIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.033902 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.034796 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BRWD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.006388 (rank : 60) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.007973 (rank : 56) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.031755 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
BAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.012472 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PU7, Q6ZQE6 | Gene names | Bap1, Kiaa0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Ubiquitin C-terminal hydrolase X4) (UCH-X4). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.035341 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CSPG4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.008656 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHY0, Q5DTG1, Q8BPI8, Q8CE79 | Gene names | Cspg4, An2, Kiaa4232, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (AN2 proteoglycan). | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.026675 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.008703 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.022677 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.007496 (rank : 59) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.043562 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.016602 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.044950 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.007705 (rank : 58) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CT166_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.072067 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H1L0 | Gene names | C20orf166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf166. | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.054105 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.055004 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.061829 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.060196 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.052661 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.056125 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
CD5R2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.5201e-129 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13319, Q4ZFW6 | Gene names | CDK5R2, NCK5AI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
CD5R2_MOUSE
|
||||||
| NC score | 0.985870 (rank : 2) | θ value | 2.51683e-135 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35926, O35277 | Gene names | Cdk5r2, Nck5ai | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
CD5R1_MOUSE
|
||||||
| NC score | 0.935963 (rank : 3) | θ value | 1.36269e-64 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61809, Q62938 | Gene names | Cdk5r1, Cdk5r, Nck5a | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
CD5R1_HUMAN
|
||||||
| NC score | 0.934390 (rank : 4) | θ value | 2.17557e-62 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15078 | Gene names | CDK5R1, CDK5R, NCK5A | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.102618 (rank : 5) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.090563 (rank : 6) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.082878 (rank : 7) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CT166_HUMAN
|
||||||
| NC score | 0.072067 (rank : 8) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H1L0 | Gene names | C20orf166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf166. | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.072039 (rank : 9) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.070787 (rank : 10) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.070373 (rank : 11) | θ value | 0.00020696 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.061829 (rank : 12) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.061301 (rank : 13) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.061248 (rank : 14) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.061102 (rank : 15) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.060196 (rank : 16) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.058615 (rank : 17) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.056125 (rank : 18) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.055004 (rank : 19) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.054105 (rank : 20) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.052797 (rank : 21) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.052661 (rank : 22) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.049239 (rank : 23) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.046373 (rank : 24) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.046086 (rank : 25) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.045947 (rank : 26) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.044983 (rank : 27) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.044950 (rank : 28) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.043898 (rank : 29) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.043562 (rank : 30) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.042645 (rank : 31) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.039585 (rank : 32) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.037346 (rank : 33) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.035341 (rank : 34) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.035196 (rank : 35) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.034796 (rank : 36) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PAIP1_HUMAN
|
||||||
| NC score | 0.033902 (rank : 37) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
EFNB3_MOUSE
|
||||||
| NC score | 0.032459 (rank : 38) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35393 | Gene names | Efnb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin-B3 precursor. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.031755 (rank : 39) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
SCAP_HUMAN
|
||||||
| NC score | 0.029965 (rank : 40) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
EFNB3_HUMAN
|
||||||
| NC score | 0.027820 (rank : 41) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15768, O00680, Q8TBH7, Q92875 | Gene names | EFNB3, EPLG8, LERK8 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin-B3 precursor (EPH-related receptor tyrosine kinase ligand 8) (LERK-8) (EPH-related receptor transmembrane ligand ELK-L3). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.026675 (rank : 42) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.022677 (rank : 43) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.022102 (rank : 44) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.020034 (rank : 45) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.017525 (rank : 46) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.016602 (rank : 47) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.016189 (rank : 48) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.014371 (rank : 49) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
TBX18_HUMAN
|
||||||
| NC score | 0.012599 (rank : 50) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
BAP1_MOUSE
|
||||||
| NC score | 0.012472 (rank : 51) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PU7, Q6ZQE6 | Gene names | Bap1, Kiaa0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Ubiquitin C-terminal hydrolase X4) (UCH-X4). | |||||
|
PP16A_HUMAN
|
||||||
| NC score | 0.011358 (rank : 52) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
ADA19_HUMAN
|
||||||
| NC score | 0.008936 (rank : 53) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.008703 (rank : 54) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
CSPG4_MOUSE
|
||||||
| NC score | 0.008656 (rank : 55) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHY0, Q5DTG1, Q8BPI8, Q8CE79 | Gene names | Cspg4, An2, Kiaa4232, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (AN2 proteoglycan). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.007973 (rank : 56) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
PUNC_MOUSE
|
||||||
| NC score | 0.007745 (rank : 57) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BQC3, O70246, Q792T2, Q9Z2S6 | Gene names | Punc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.007705 (rank : 58) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.007496 (rank : 59) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
BRWD1_HUMAN
|
||||||
| NC score | 0.006388 (rank : 60) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||