Please be patient as the page loads
|
KCNN2_HUMAN
|
||||||
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCNN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.988177 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989156 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998924 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.978393 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.983771 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
KCNN4_MOUSE
|
||||||
| θ value | 1.35324e-88 (rank : 7) | NC score | 0.955595 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNN4_HUMAN
|
||||||
| θ value | 3.68522e-86 (rank : 8) | NC score | 0.953514 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 9) | NC score | 0.360774 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 10) | NC score | 0.365699 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 11) | NC score | 0.305245 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 12) | NC score | 0.305263 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 13) | NC score | 0.344538 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 14) | NC score | 0.342125 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 15) | NC score | 0.305058 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 16) | NC score | 0.305225 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 17) | NC score | 0.302769 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 18) | NC score | 0.300929 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 19) | NC score | 0.301062 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 20) | NC score | 0.301809 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 21) | NC score | 0.335462 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 22) | NC score | 0.332378 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 23) | NC score | 0.298379 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 24) | NC score | 0.342824 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 25) | NC score | 0.341600 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 26) | NC score | 0.293177 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 27) | NC score | 0.295778 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 28) | NC score | 0.288071 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 29) | NC score | 0.288918 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 30) | NC score | 0.288271 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 31) | NC score | 0.286570 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 32) | NC score | 0.286980 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 33) | NC score | 0.286370 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 34) | NC score | 0.280400 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 35) | NC score | 0.280441 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 36) | NC score | 0.286400 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 37) | NC score | 0.281929 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 38) | NC score | 0.281680 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 39) | NC score | 0.265915 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 40) | NC score | 0.266637 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 41) | NC score | 0.281200 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 42) | NC score | 0.273014 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 43) | NC score | 0.272800 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 44) | NC score | 0.280270 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNK1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 45) | NC score | 0.135082 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 46) | NC score | 0.266554 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 47) | NC score | 0.218041 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 48) | NC score | 0.218435 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 49) | NC score | 0.258454 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 50) | NC score | 0.258518 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.258265 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
ANGL7_MOUSE
|
||||||
| θ value | 0.279714 (rank : 52) | NC score | 0.032076 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R1Q3 | Gene names | Angptl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | 0.255077 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.255253 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNK1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.120134 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.133975 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.029766 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
ANR35_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.018246 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.024327 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.023323 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.263414 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.269260 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
NOL10_MOUSE
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.043930 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5RJG1 | Gene names | Nol10, Gm67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.023342 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
ADCY6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.016889 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43306, Q9NR75 | Gene names | ADCY6, KIAA0422 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.022894 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.019662 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.259886 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNK9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.110937 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.110555 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.256577 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
INOC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.019095 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
INOC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.019601 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.255137 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
AAT1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.049480 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z4T9, Q68DX2, Q8TD41, Q96A45, Q96JE8 | Gene names | AAT1, C3orf15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMY-1-associating protein expressed in testis 1 (AAT-1). | |||||
|
DC1L2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.018020 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.038828 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.019656 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.021859 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
A16L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.014514 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q676U5, Q6IPN1, Q6UXW4, Q6ZVZ5, Q8NCY2, Q96JV5, Q9H619 | Gene names | ATG16L1, APG16L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
CKAP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.020227 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.025189 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.019968 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.010776 (rank : 128) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.017761 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
ADCY6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.014336 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01341 | Gene names | Adcy6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.020988 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
NOL10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.035336 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BSC4, Q53RC9, Q96TA5, Q9H7Y7, Q9H855 | Gene names | NOL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.006470 (rank : 131) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.018016 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.113416 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.114106 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.092854 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
A16L1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.012677 (rank : 124) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C0J2, Q6KAT7, Q80U97, Q80U98, Q80U99, Q80Y53, Q9DB63 | Gene names | Atg16l1, Apg16l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.016937 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
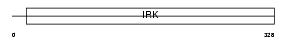
IRK13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.009523 (rank : 129) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60928, O76023, Q8N3Y4 | Gene names | KCNJ13 | |||
|
Domain Architecture |
|
|||||
| Description | Inward rectifier potassium channel 13 (Potassium channel, inwardly rectifying subfamily J member 13) (Inward rectifier K(+) channel Kir7.1). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.028771 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.099704 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.099552 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNKD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.066347 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.117307 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.012253 (rank : 125) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.011902 (rank : 126) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.018222 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
DCDC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.006407 (rank : 132) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.013669 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.021593 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.021575 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNKD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.059204 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.020224 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.009412 (rank : 130) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.011406 (rank : 127) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GCC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.024788 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.014781 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.013653 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.091018 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.015490 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
KCD15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.056365 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96SI1, Q9BVI6 | Gene names | KCTD15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCD15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.054077 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K0E1 | Gene names | Kctd15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.071908 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.076225 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.055707 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.076468 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.052404 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.085292 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCTD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.057876 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q719H9 | Gene names | KCTD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.057876 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5M956, Q5EER9 | Gene names | Kctd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.052424 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WVF5 | Gene names | KCTD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.052713 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D7X1, Q8CCQ3, Q9CYK4 | Gene names | Kctd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.057214 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NC69, Q8NBS6, Q8TCA6 | Gene names | KCTD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.057667 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BNL5, Q3TXX1 | Gene names | Kctd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.050584 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BJK1, Q3USA6, Q8C0S7 | Gene names | Kctd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
KCNN2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.998924 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN1_MOUSE
|
||||||
| NC score | 0.989156 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_HUMAN
|
||||||
| NC score | 0.988177 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.983771 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.978393 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN4_MOUSE
|
||||||
| NC score | 0.955595 (rank : 7) | θ value | 1.35324e-88 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNN4_HUMAN
|
||||||
| NC score | 0.953514 (rank : 8) | θ value | 3.68522e-86 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.365699 (rank : 9) | θ value | 0.000121331 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.360774 (rank : 10) | θ value | 2.44474e-05 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.344538 (rank : 11) | θ value | 0.00020696 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.342824 (rank : 12) | θ value | 0.00509761 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.342125 (rank : 13) | θ value | 0.000270298 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.341600 (rank : 14) | θ value | 0.00509761 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.335462 (rank : 15) | θ value | 0.00175202 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.332378 (rank : 16) | θ value | 0.00228821 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.305263 (rank : 17) | θ value | 0.00020696 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.305245 (rank : 18) | θ value | 0.00020696 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.305225 (rank : 19) | θ value | 0.00035302 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.305058 (rank : 20) | θ value | 0.00035302 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.302769 (rank : 21) | θ value | 0.000602161 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.301809 (rank : 22) | θ value | 0.000786445 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.301062 (rank : 23) | θ value | 0.000786445 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.300929 (rank : 24) | θ value | 0.000786445 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.298379 (rank : 25) | θ value | 0.00390308 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.295778 (rank : 26) | θ value | 0.00665767 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.293177 (rank : 27) | θ value | 0.00665767 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.288918 (rank : 28) | θ value | 0.0113563 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.288271 (rank : 29) | θ value | 0.0148317 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.288071 (rank : 30) | θ value | 0.00665767 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.286980 (rank : 31) | θ value | 0.0193708 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.286570 (rank : 32) | θ value | 0.0193708 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.286400 (rank : 33) | θ value | 0.0330416 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.286370 (rank : 34) | θ value | 0.0252991 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.281929 (rank : 35) | θ value | 0.0431538 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.281680 (rank : 36) | θ value | 0.0431538 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.281200 (rank : 37) | θ value | 0.0736092 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.280441 (rank : 38) | θ value | 0.0252991 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.280400 (rank : 39) | θ value | 0.0252991 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.280270 (rank : 40) | θ value | 0.0961366 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.273014 (rank : 41) | θ value | 0.0736092 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.272800 (rank : 42) | θ value | 0.0736092 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.269260 (rank : 43) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.266637 (rank : 44) | θ value | 0.0431538 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.266554 (rank : 45) | θ value | 0.125558 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.265915 (rank : 46) | θ value | 0.0431538 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.263414 (rank : 47) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.259886 (rank : 48) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.258518 (rank : 49) | θ value | 0.21417 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.258454 (rank : 50) | θ value | 0.21417 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.258265 (rank : 51) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.256577 (rank : 52) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.255253 (rank : 53) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.255137 (rank : 54) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.255077 (rank : 55) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.218435 (rank : 56) | θ value | 0.21417 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.218041 (rank : 57) | θ value | 0.21417 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCNK1_HUMAN
|
||||||
| NC score | 0.135082 (rank : 58) | θ value | 0.125558 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.133975 (rank : 59) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK1_MOUSE
|
||||||
| NC score | 0.120134 (rank : 60) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.117307 (rank : 61) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.114106 (rank : 62) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.113416 (rank : 63) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNK9_HUMAN
|
||||||
| NC score | 0.110937 (rank : 64) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.110555 (rank : 65) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.099704 (rank : 66) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.099552 (rank : 67) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNT1_HUMAN
|
||||||
| NC score | 0.092854 (rank : 68) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.091018 (rank : 69) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.085292 (rank : 70) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.076468 (rank : 71) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.076225 (rank : 72) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.071908 (rank : 73) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNKD_HUMAN
|
||||||
| NC score | 0.066347 (rank : 74) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKD_MOUSE
|
||||||
| NC score | 0.059204 (rank : 75) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCTD1_HUMAN
|
||||||
| NC score | 0.057876 (rank : 76) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q719H9 | Gene names | KCTD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD1_MOUSE
|
||||||
| NC score | 0.057876 (rank : 77) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5M956, Q5EER9 | Gene names | Kctd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD6_MOUSE
|
||||||
| NC score | 0.057667 (rank : 78) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BNL5, Q3TXX1 | Gene names | Kctd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD6_HUMAN
|
||||||
| NC score | 0.057214 (rank : 79) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NC69, Q8NBS6, Q8TCA6 | Gene names | KCTD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCD15_HUMAN
|
||||||
| NC score | 0.056365 (rank : 80) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96SI1, Q9BVI6 | Gene names | KCTD15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.055707 (rank : 81) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCD15_MOUSE
|
||||||
| NC score | 0.054077 (rank : 82) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K0E1 | Gene names | Kctd15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCTD4_MOUSE
|
||||||
| NC score | 0.052713 (rank : 83) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D7X1, Q8CCQ3, Q9CYK4 | Gene names | Kctd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD4_HUMAN
|
||||||
| NC score | 0.052424 (rank : 84) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WVF5 | Gene names | KCTD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.052404 (rank : 85) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCTD7_MOUSE
|
||||||
| NC score | 0.050584 (rank : 86) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BJK1, Q3USA6, Q8C0S7 | Gene names | Kctd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
AAT1_HUMAN
|
||||||
| NC score | 0.049480 (rank : 87) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z4T9, Q68DX2, Q8TD41, Q96A45, Q96JE8 | Gene names | AAT1, C3orf15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMY-1-associating protein expressed in testis 1 (AAT-1). | |||||
|
NOL10_MOUSE
|
||||||
| NC score | 0.043930 (rank : 88) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5RJG1 | Gene names | Nol10, Gm67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.038828 (rank : 89) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
NOL10_HUMAN
|
||||||
| NC score | 0.035336 (rank : 90) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BSC4, Q53RC9, Q96TA5, Q9H7Y7, Q9H855 | Gene names | NOL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
ANGL7_MOUSE
|
||||||
| NC score | 0.032076 (rank : 91) | θ value | 0.279714 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R1Q3 | Gene names | Angptl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.029766 (rank : 92) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.028771 (rank : 93) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.025189 (rank : 94) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
GCC1_HUMAN
|
||||||
| NC score | 0.024788 (rank : 95) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.024327 (rank : 96) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.023342 (rank : 97) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.023323 (rank : 98) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.022894 (rank : 99) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.021859 (rank : 100) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.021593 (rank : 101) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.021575 (rank : 102) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.020988 (rank : 103) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
CKAP2_MOUSE
|
||||||
| NC score | 0.020227 (rank : 104) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.020224 (rank : 105) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.019968 (rank : 106) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.019662 (rank : 107) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.019656 (rank : 108) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
INOC1_MOUSE
|
||||||
| NC score | 0.019601 (rank : 109) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
INOC1_HUMAN
|
||||||
| NC score | 0.019095 (rank : 110) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.018246 (rank : 111) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.018222 (rank : 112) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
DC1L2_MOUSE
|
||||||
| NC score | 0.018020 (rank : 113) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.018016 (rank : 114) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.017761 (rank : 115) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.016937 (rank : 116) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
ADCY6_HUMAN
|
||||||
| NC score | 0.016889 (rank : 117) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43306, Q9NR75 | Gene names | ADCY6, KIAA0422 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.015490 (rank : 118) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.014781 (rank : 119) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
A16L1_HUMAN
|
||||||
| NC score | 0.014514 (rank : 120) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q676U5, Q6IPN1, Q6UXW4, Q6ZVZ5, Q8NCY2, Q96JV5, Q9H619 | Gene names | ATG16L1, APG16L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
ADCY6_MOUSE
|
||||||
| NC score | 0.014336 (rank : 121) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01341 | Gene names | Adcy6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.013669 (rank : 122) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.013653 (rank : 123) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
A16L1_MOUSE
|
||||||
| NC score | 0.012677 (rank : 124) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C0J2, Q6KAT7, Q80U97, Q80U98, Q80U99, Q80Y53, Q9DB63 | Gene names | Atg16l1, Apg16l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.012253 (rank : 125) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.011902 (rank : 126) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.011406 (rank : 127) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.010776 (rank : 128) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
IRK13_HUMAN
|
||||||
| NC score | 0.009523 (rank : 129) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60928, O76023, Q8N3Y4 | Gene names | KCNJ13 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 13 (Potassium channel, inwardly rectifying subfamily J member 13) (Inward rectifier K(+) channel Kir7.1). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.009412 (rank : 130) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.006470 (rank : 131) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
DCDC2_MOUSE
|
||||||
| NC score | 0.006407 (rank : 132) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||