Please be patient as the page loads
|
ANGL7_MOUSE
|
||||||
| SwissProt Accessions | Q8R1Q3 | Gene names | Angptl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ANGL7_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R1Q3 | Gene names | Angptl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7). | |||||
|
ANGL7_HUMAN
|
||||||
| θ value | 8.28183e-171 (rank : 2) | NC score | 0.997612 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O43827, Q4ZGK4 | Gene names | ANGPTL7, CDT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7) (Angiopoietin-like factor) (Cornea-derived transcript 6 protein). | |||||
|
ANGP4_HUMAN
|
||||||
| θ value | 2.10721e-57 (rank : 3) | NC score | 0.956215 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y264, Q9H4Z4 | Gene names | ANGPT4, ANG3, ANG4 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-4 precursor (ANG-4) (ANG-3). | |||||
|
ANGP1_MOUSE
|
||||||
| θ value | 1.97229e-55 (rank : 4) | NC score | 0.942851 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O08538 | Gene names | Angpt1, Agpt | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
ANGP2_HUMAN
|
||||||
| θ value | 2.57589e-55 (rank : 5) | NC score | 0.915215 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15123, Q9NRR7, Q9P2Y7 | Gene names | ANGPT2 | |||
|
Domain Architecture |
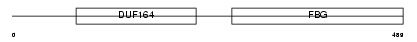
|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
ANGP2_MOUSE
|
||||||
| θ value | 3.36421e-55 (rank : 6) | NC score | 0.911166 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
ANGP1_HUMAN
|
||||||
| θ value | 7.49467e-55 (rank : 7) | NC score | 0.942277 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15389 | Gene names | ANGPT1, KIAA0003 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
ANGL1_HUMAN
|
||||||
| θ value | 1.08223e-53 (rank : 8) | NC score | 0.965970 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95841, Q5T5Z5 | Gene names | ANGPTL1, ANG3, ANGPT3, ARP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 1 precursor (Angiopoietin-like 1) (Angiopoietin-3) (ANG-3). | |||||
|
ANGL1_MOUSE
|
||||||
| θ value | 1.84601e-53 (rank : 9) | NC score | 0.964630 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q640P2, Q3UQL1, Q9CZ81 | Gene names | Angptl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 1 precursor (Angiopoietin-like 1). | |||||
|
ANGP4_MOUSE
|
||||||
| θ value | 4.11246e-53 (rank : 10) | NC score | 0.947492 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WVH6 | Gene names | Angpt4, Agpt4, Ang3 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-4 precursor (ANG-4) (ANG-3). | |||||
|
FCN1_HUMAN
|
||||||
| θ value | 1.72781e-51 (rank : 11) | NC score | 0.903039 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00602, Q92596 | Gene names | FCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Ficolin-1 precursor (Collagen/fibrinogen domain-containing protein 1) (Ficolin-A) (Ficolin A) (M-Ficolin). | |||||
|
FGL1_HUMAN
|
||||||
| θ value | 8.57503e-51 (rank : 12) | NC score | 0.982531 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08830, Q4PJH9, Q96KW6, Q96QM6 | Gene names | FGL1, HFREP1 | |||
|
Domain Architecture |
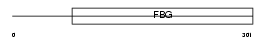
|
|||||
| Description | Fibrinogen-like protein 1 precursor (Hepatocyte-derived fibrinogen- related protein 1) (HFREP-1) (Hepassocin) (HP-041). | |||||
|
ANGL2_MOUSE
|
||||||
| θ value | 1.91031e-50 (rank : 13) | NC score | 0.963117 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9R045 | Gene names | Angptl2, Arp2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 2 precursor (Angiopoietin-like 2). | |||||
|
ANGL2_HUMAN
|
||||||
| θ value | 2.49495e-50 (rank : 14) | NC score | 0.962816 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKU9, Q5JT58 | Gene names | ANGPTL2, ARP2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 2 precursor (Angiopoietin-like 2). | |||||
|
FCN2_HUMAN
|
||||||
| θ value | 3.60269e-49 (rank : 15) | NC score | 0.929025 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15485, Q9UC57 | Gene names | FCN2 | |||
|
Domain Architecture |

|
|||||
| Description | Ficolin-2 precursor (Collagen/fibrinogen domain-containing protein 2) (Ficolin-B) (Ficolin B) (Serum lectin p35) (EBP-37) (Hucolin) (L- Ficolin). | |||||
|
MFAP4_HUMAN
|
||||||
| θ value | 7.51208e-47 (rank : 16) | NC score | 0.978751 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P55083, Q6P680 | Gene names | MFAP4 | |||
|
Domain Architecture |
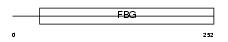
|
|||||
| Description | Microfibril-associated glycoprotein 4 precursor. | |||||
|
ANGL6_HUMAN
|
||||||
| θ value | 9.81109e-47 (rank : 17) | NC score | 0.969712 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NI99, Q9BZZ0 | Gene names | ANGPTL6, AGF, ARP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 6 precursor (Angiopoietin-like 6) (Angiopoietin-related growth factor) (Angiopoietin-related protein 5). | |||||
|
ANGL6_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 18) | NC score | 0.970886 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R0Z6 | Gene names | Angptl6, Agf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 6 precursor (Angiopoietin-like 6) (Angiopoietin-related growth factor). | |||||
|
MFAP4_MOUSE
|
||||||
| θ value | 2.85459e-46 (rank : 19) | NC score | 0.978678 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D1H9, Q5NCN1 | Gene names | Mfap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microfibril-associated glycoprotein 4 precursor. | |||||
|
FIBA_HUMAN
|
||||||
| θ value | 4.86918e-46 (rank : 20) | NC score | 0.962954 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P02671, Q9BX62, Q9UCH2 | Gene names | FGA | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A]. | |||||
|
TENR_HUMAN
|
||||||
| θ value | 4.86918e-46 (rank : 21) | NC score | 0.668280 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
FGL2_MOUSE
|
||||||
| θ value | 8.30557e-46 (rank : 22) | NC score | 0.966980 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12804 | Gene names | Fgl2, Fiblp | |||
|
Domain Architecture |
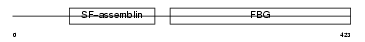
|
|||||
| Description | Fibroleukin precursor (Fibrinogen-like protein 2) (Prothrombinase) (Cytotoxic T-lymphocyte-specific protein). | |||||
|
FGL2_HUMAN
|
||||||
| θ value | 1.41672e-45 (rank : 23) | NC score | 0.966364 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14314 | Gene names | FGL2 | |||
|
Domain Architecture |
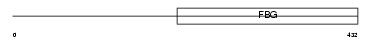
|
|||||
| Description | Fibroleukin precursor (Fibrinogen-like protein 2) (pT49). | |||||
|
FCN1_MOUSE
|
||||||
| θ value | 3.15612e-45 (rank : 24) | NC score | 0.879141 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O70165 | Gene names | Fcn1, Fcna | |||
|
Domain Architecture |

|
|||||
| Description | Ficolin-1 precursor (Collagen/fibrinogen domain-containing protein 1) (Ficolin-A) (Ficolin A) (M-Ficolin). | |||||
|
TENR_MOUSE
|
||||||
| θ value | 3.15612e-45 (rank : 25) | NC score | 0.666020 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
TENN_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 26) | NC score | 0.710738 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQP3, Q5R360 | Gene names | TNN | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
FCN3_HUMAN
|
||||||
| θ value | 8.59492e-43 (rank : 27) | NC score | 0.945994 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75636, Q6IBJ5, Q8WW86 | Gene names | FCN3, HAKA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ficolin-3 precursor (Collagen/fibrinogen domain-containing protein 3) (Collagen/fibrinogen domain-containing lectin 3 p35) (Hakata antigen). | |||||
|
TENN_MOUSE
|
||||||
| θ value | 3.26607e-42 (rank : 28) | NC score | 0.680228 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80Z71 | Gene names | Tnn | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
FIBB_HUMAN
|
||||||
| θ value | 4.2656e-42 (rank : 29) | NC score | 0.957293 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02675 | Gene names | FGB | |||
|
Domain Architecture |
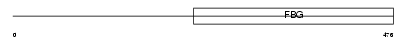
|
|||||
| Description | Fibrinogen beta chain precursor [Contains: Fibrinopeptide B]. | |||||
|
TENX_HUMAN
|
||||||
| θ value | 9.50281e-42 (rank : 30) | NC score | 0.541489 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
TENA_HUMAN
|
||||||
| θ value | 1.62093e-41 (rank : 31) | NC score | 0.540378 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TENA_MOUSE
|
||||||
| θ value | 1.62093e-41 (rank : 32) | NC score | 0.554144 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
FIBB_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 33) | NC score | 0.957573 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K0E8, Q91ZP1 | Gene names | Fgb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrinogen beta chain precursor [Contains: Fibrinopeptide B]. | |||||
|
ANGL4_HUMAN
|
||||||
| θ value | 3.73686e-38 (rank : 34) | NC score | 0.960617 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BY76, Q53HQ6, Q53HU1, Q6UXN0, Q9HBV4, Q9NZU4, Q9Y5B3 | Gene names | ANGPTL4, ARP4, HFARP, PGAR | |||
|
Domain Architecture |
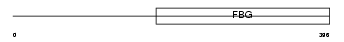
|
|||||
| Description | Angiopoietin-related protein 4 precursor (Angiopoietin-like 4) (Hepatic fibrinogen/angiopoietin-related protein) (HFARP). | |||||
|
FIBG_HUMAN
|
||||||
| θ value | 8.32485e-38 (rank : 35) | NC score | 0.955274 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P02679, P04469, P04470, Q96A14, Q96KJ3 | Gene names | FGG | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen gamma chain precursor. | |||||
|
ANGL4_MOUSE
|
||||||
| θ value | 9.20422e-37 (rank : 36) | NC score | 0.959399 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z1P8, Q78ZJ9, Q9JHX7, Q9JLX7 | Gene names | Angptl4, Farp, Fiaf, Ng27 | |||
|
Domain Architecture |
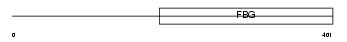
|
|||||
| Description | Angiopoietin-related protein 4 precursor (Angiopoietin-like 4) (Hepatic fibrinogen/angiopoietin-related protein) (HFARP) (Fasting- induced adipose factor) (Secreted protein Bk89) (425O18-1). | |||||
|
FIBG_MOUSE
|
||||||
| θ value | 4.568e-36 (rank : 37) | NC score | 0.958009 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8VCM7, Q8WUR3, Q91ZP0 | Gene names | Fgg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrinogen gamma chain precursor. | |||||
|
ANGL3_MOUSE
|
||||||
| θ value | 7.29293e-34 (rank : 38) | NC score | 0.911353 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9R182 | Gene names | Angptl3 | |||
|
Domain Architecture |
|
|||||
| Description | Angiopoietin-related protein 3 precursor (Angiopoietin-like 3). | |||||
|
ANGL3_HUMAN
|
||||||
| θ value | 3.61944e-33 (rank : 39) | NC score | 0.916821 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y5C1 | Gene names | ANGPTL3, ANGPT5 | |||
|
Domain Architecture |
|
|||||
| Description | Angiopoietin-related protein 3 precursor (Angiopoietin-like 3) (Angiopoietin-5) (ANG-5). | |||||
|
ANGL5_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 40) | NC score | 0.962217 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86XS5, Q86VR9 | Gene names | ANGPTL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 5 precursor (Angiopoietin-like 5). | |||||
|
KCNN1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 41) | NC score | 0.036027 (rank : 139) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.033426 (rank : 140) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
BCAS2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.044931 (rank : 137) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75934, Q6FGS0 | Gene names | BCAS2, DAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 (DNA amplified in mammary carcinoma 1 protein) (Spliceosome-associated protein SPF 27). | |||||
|
BCAS2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.044931 (rank : 138) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D287, Q3TKJ5, Q91YX8, Q9D2U4, Q9DAI0 | Gene names | Bcas2, Dam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 homolog (DNA amplified in mammary carcinoma 1 protein). | |||||
|
KCNN2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.032076 (rank : 141) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.031945 (rank : 142) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.027307 (rank : 145) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.022859 (rank : 147) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.012159 (rank : 163) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.017690 (rank : 153) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
ITL1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.150229 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88310 | Gene names | Itln1a, Intl, Itln, Itlna | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intelectin-1a precursor (Intestinal lactoferrin receptor) (Galactofuranose-binding lectin). | |||||
|
ITL1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.153531 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80ZA0 | Gene names | Itln1b, Itln2, Itlnb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intelectin-1b precursor (Intelectin-2). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.017103 (rank : 154) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.019270 (rank : 151) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.019153 (rank : 152) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
HDAC5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.005391 (rank : 170) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.016317 (rank : 157) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.019858 (rank : 150) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
IDE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.010881 (rank : 165) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.031085 (rank : 143) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
LMNA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.007104 (rank : 168) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 653 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02545, P02546, Q969I8, Q96JA2 | Gene names | LMNA, LMN1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C (70 kDa lamin) (NY-REN-32 antigen). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.029715 (rank : 144) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HUS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.008623 (rank : 167) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BQY8, O70543, Q6P8H5 | Gene names | Hus1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Checkpoint protein HUS1 (mHUS1). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.014776 (rank : 161) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
ST18_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.011131 (rank : 164) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
ITLN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.095039 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WWA0, Q6YDJ3, Q9NP67 | Gene names | ITLN1, INTL, ITLN, LFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intelectin-1 precursor (Intestinal lactoferrin receptor) (Galactofuranose-binding lectin) (Endothelial lectin HL-1) (Omentin). | |||||
|
ITLN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.125173 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WWU7 | Gene names | ITLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intelectin-2 precursor (Endothelial lectin HL-2). | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.014627 (rank : 162) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LMNA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.006370 (rank : 169) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48678, P11516, P97859, Q3TIH0, Q3TTS8, Q3UCA0, Q3UCJ8, Q91WF2, Q9DC21 | Gene names | Lmna, Lmn1 | |||
|
Domain Architecture |
|
|||||
| Description | Lamin-A/C. | |||||
|
T3JAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.020120 (rank : 149) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
PLXD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.004257 (rank : 172) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.016185 (rank : 158) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.026474 (rank : 146) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CHGUT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.003863 (rank : 173) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.016383 (rank : 156) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.016986 (rank : 155) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.015376 (rank : 160) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PLCE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.004954 (rank : 171) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D1E8, Q3U702, Q8BG61, Q8CGN6 | Gene names | Agpat5, D8Ertd319e | |||
|
Domain Architecture |
|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase epsilon (EC 2.3.1.51) (1-AGP acyltransferase 5) (1-AGPAT 5) (Lysophosphatidic acid acyltransferase-epsilon) (LPAAT-epsilon) (1-acylglycerol-3-phosphate O-acyltransferase 5). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.020317 (rank : 148) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.009883 (rank : 166) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.016111 (rank : 159) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.105390 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
COCA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.124888 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COCA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.127272 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COEA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.121036 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
COEA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.117800 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
COKA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.119532 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
DLK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.064031 (rank : 101) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.061917 (rank : 105) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.052749 (rank : 134) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.052790 (rank : 133) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.056611 (rank : 120) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
DLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.056275 (rank : 121) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
DLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.055968 (rank : 123) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |
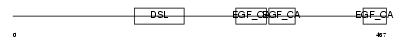
|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.055563 (rank : 124) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DNER_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.057580 (rank : 119) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
DNER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.057907 (rank : 117) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZM4, Q3U697, Q8R226, Q8R4T6, Q8VD97 | Gene names | Dner, Bet, Bret | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor (Brain EGF repeat-containing transmembrane protein). | |||||
|
EGFL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.065568 (rank : 100) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
EGFL9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.063553 (rank : 102) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
FINC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.171966 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.167585 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
FNDC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.058960 (rank : 112) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2H6, Q5JVF9, Q6EVH3, Q6N020, Q6P9D5, Q9H1W1 | Gene names | FNDC3A, FNDC3, KIAA0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
FNDC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.071810 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX90, Q2VEY7, Q6ZQ15, Q811D3, Q8BTM3 | Gene names | Fndc3a, D14Ertd453e, Fndc3, Kiaa0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
HHIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.067187 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QV1, Q6PK09, Q8NCI7, Q9BXK3, Q9H1J4, Q9H7E7 | Gene names | HHIP, HIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
HHIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.067553 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TN16, Q8C0B0, Q9WU59 | Gene names | Hhip, Hip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
ITB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.069123 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.070132 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.074048 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05107, Q16418 | Gene names | ITGB2, CD18 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/p150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
ITB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.073309 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
ITB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.075650 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05106, O15495, Q13413, Q14648, Q16499 | Gene names | ITGB3, GP3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
ITB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.075938 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54890 | Gene names | Itgb3 | |||
|
Domain Architecture |
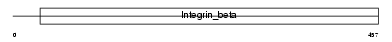
|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
ITB4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.069096 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
ITB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.073019 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18084 | Gene names | ITGB5 | |||
|
Domain Architecture |
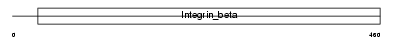
|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
ITB5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.075611 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |
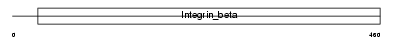
|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
ITB6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.072747 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18564, Q16500 | Gene names | ITGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ITB6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.070964 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ITB7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.070715 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26010 | Gene names | ITGB7 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-7 precursor. | |||||
|
ITB7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.067355 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26011, Q3U2M1, Q64656 | Gene names | Itgb7 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-7 precursor (Integrin beta-P) (M290 IEL antigen). | |||||
|
ITB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.077349 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
JAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.059371 (rank : 111) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.058875 (rank : 113) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
JAG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.058406 (rank : 115) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.058130 (rank : 116) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
KR101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.056022 (rank : 122) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR102_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.067147 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR104_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.066501 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR106_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.060136 (rank : 109) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR107_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.058531 (rank : 114) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR108_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.053141 (rank : 132) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.054342 (rank : 127) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.061939 (rank : 103) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR171_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.142252 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KR412_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.069247 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
KR414_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054444 (rank : 126) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KR510_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.065751 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KR511_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.096902 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.079888 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
KRA13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.068607 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KRA42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.061932 (rank : 104) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
KRA44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.086049 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KRA45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.079422 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.071770 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.095325 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.088675 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.096944 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.098422 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.086566 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.103393 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.110635 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.102747 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.090820 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA92_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.068381 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.054107 (rank : 128) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA99_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.059533 (rank : 110) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.060832 (rank : 108) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
NAGPA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.113543 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UK23, Q96EJ8 | Gene names | NAGPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NAGPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.112538 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
PTPRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.101130 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
PTPRJ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.057868 (rank : 118) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTPRJ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.053293 (rank : 131) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.075276 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
RELN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.060902 (rank : 107) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |
|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
RELN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.061188 (rank : 106) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60841, Q9CUA6 | Gene names | Reln, Rl | |||
|
Domain Architecture |
|
|||||
| Description | Reelin precursor (EC 3.4.21.-) (Reeler protein). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.072850 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.072594 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
SREC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.068190 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
STAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.052267 (rank : 135) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054031 (rank : 129) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.054687 (rank : 125) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.053505 (rank : 130) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
USH2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.050206 (rank : 136) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
WIF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.105461 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
WIF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.103538 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
ANGL7_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R1Q3 | Gene names | Angptl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7). | |||||
|
ANGL7_HUMAN
|
||||||
| NC score | 0.997612 (rank : 2) | θ value | 8.28183e-171 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O43827, Q4ZGK4 | Gene names | ANGPTL7, CDT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7) (Angiopoietin-like factor) (Cornea-derived transcript 6 protein). | |||||
|
FGL1_HUMAN
|
||||||
| NC score | 0.982531 (rank : 3) | θ value | 8.57503e-51 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08830, Q4PJH9, Q96KW6, Q96QM6 | Gene names | FGL1, HFREP1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen-like protein 1 precursor (Hepatocyte-derived fibrinogen- related protein 1) (HFREP-1) (Hepassocin) (HP-041). | |||||
|
MFAP4_HUMAN
|
||||||
| NC score | 0.978751 (rank : 4) | θ value | 7.51208e-47 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P55083, Q6P680 | Gene names | MFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microfibril-associated glycoprotein 4 precursor. | |||||
|
MFAP4_MOUSE
|
||||||
| NC score | 0.978678 (rank : 5) | θ value | 2.85459e-46 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D1H9, Q5NCN1 | Gene names | Mfap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microfibril-associated glycoprotein 4 precursor. | |||||
|
ANGL6_MOUSE
|
||||||
| NC score | 0.970886 (rank : 6) | θ value | 1.28137e-46 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R0Z6 | Gene names | Angptl6, Agf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 6 precursor (Angiopoietin-like 6) (Angiopoietin-related growth factor). | |||||
|
ANGL6_HUMAN
|
||||||
| NC score | 0.969712 (rank : 7) | θ value | 9.81109e-47 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NI99, Q9BZZ0 | Gene names | ANGPTL6, AGF, ARP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 6 precursor (Angiopoietin-like 6) (Angiopoietin-related growth factor) (Angiopoietin-related protein 5). | |||||
|
FGL2_MOUSE
|
||||||
| NC score | 0.966980 (rank : 8) | θ value | 8.30557e-46 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12804 | Gene names | Fgl2, Fiblp | |||
|
Domain Architecture |

|
|||||
| Description | Fibroleukin precursor (Fibrinogen-like protein 2) (Prothrombinase) (Cytotoxic T-lymphocyte-specific protein). | |||||
|
FGL2_HUMAN
|
||||||
| NC score | 0.966364 (rank : 9) | θ value | 1.41672e-45 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14314 | Gene names | FGL2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibroleukin precursor (Fibrinogen-like protein 2) (pT49). | |||||
|
ANGL1_HUMAN
|
||||||
| NC score | 0.965970 (rank : 10) | θ value | 1.08223e-53 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95841, Q5T5Z5 | Gene names | ANGPTL1, ANG3, ANGPT3, ARP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 1 precursor (Angiopoietin-like 1) (Angiopoietin-3) (ANG-3). | |||||
|
ANGL1_MOUSE
|
||||||
| NC score | 0.964630 (rank : 11) | θ value | 1.84601e-53 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q640P2, Q3UQL1, Q9CZ81 | Gene names | Angptl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 1 precursor (Angiopoietin-like 1). | |||||
|
ANGL2_MOUSE
|
||||||
| NC score | 0.963117 (rank : 12) | θ value | 1.91031e-50 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9R045 | Gene names | Angptl2, Arp2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 2 precursor (Angiopoietin-like 2). | |||||
|
FIBA_HUMAN
|
||||||
| NC score | 0.962954 (rank : 13) | θ value | 4.86918e-46 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P02671, Q9BX62, Q9UCH2 | Gene names | FGA | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A]. | |||||
|
ANGL2_HUMAN
|
||||||
| NC score | 0.962816 (rank : 14) | θ value | 2.49495e-50 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKU9, Q5JT58 | Gene names | ANGPTL2, ARP2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 2 precursor (Angiopoietin-like 2). | |||||
|
ANGL5_HUMAN
|
||||||
| NC score | 0.962217 (rank : 15) | θ value | 3.74554e-30 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86XS5, Q86VR9 | Gene names | ANGPTL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 5 precursor (Angiopoietin-like 5). | |||||
|
ANGL4_HUMAN
|
||||||
| NC score | 0.960617 (rank : 16) | θ value | 3.73686e-38 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BY76, Q53HQ6, Q53HU1, Q6UXN0, Q9HBV4, Q9NZU4, Q9Y5B3 | Gene names | ANGPTL4, ARP4, HFARP, PGAR | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 4 precursor (Angiopoietin-like 4) (Hepatic fibrinogen/angiopoietin-related protein) (HFARP). | |||||
|
ANGL4_MOUSE
|
||||||
| NC score | 0.959399 (rank : 17) | θ value | 9.20422e-37 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z1P8, Q78ZJ9, Q9JHX7, Q9JLX7 | Gene names | Angptl4, Farp, Fiaf, Ng27 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 4 precursor (Angiopoietin-like 4) (Hepatic fibrinogen/angiopoietin-related protein) (HFARP) (Fasting- induced adipose factor) (Secreted protein Bk89) (425O18-1). | |||||
|
FIBG_MOUSE
|
||||||
| NC score | 0.958009 (rank : 18) | θ value | 4.568e-36 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8VCM7, Q8WUR3, Q91ZP0 | Gene names | Fgg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrinogen gamma chain precursor. | |||||
|
FIBB_MOUSE
|
||||||
| NC score | 0.957573 (rank : 19) | θ value | 1.05066e-40 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K0E8, Q91ZP1 | Gene names | Fgb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrinogen beta chain precursor [Contains: Fibrinopeptide B]. | |||||
|
FIBB_HUMAN
|
||||||
| NC score | 0.957293 (rank : 20) | θ value | 4.2656e-42 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02675 | Gene names | FGB | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen beta chain precursor [Contains: Fibrinopeptide B]. | |||||
|
ANGP4_HUMAN
|
||||||
| NC score | 0.956215 (rank : 21) | θ value | 2.10721e-57 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y264, Q9H4Z4 | Gene names | ANGPT4, ANG3, ANG4 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-4 precursor (ANG-4) (ANG-3). | |||||
|
FIBG_HUMAN
|
||||||
| NC score | 0.955274 (rank : 22) | θ value | 8.32485e-38 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P02679, P04469, P04470, Q96A14, Q96KJ3 | Gene names | FGG | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen gamma chain precursor. | |||||
|
ANGP4_MOUSE
|
||||||
| NC score | 0.947492 (rank : 23) | θ value | 4.11246e-53 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WVH6 | Gene names | Angpt4, Agpt4, Ang3 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-4 precursor (ANG-4) (ANG-3). | |||||
|
FCN3_HUMAN
|
||||||
| NC score | 0.945994 (rank : 24) | θ value | 8.59492e-43 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75636, Q6IBJ5, Q8WW86 | Gene names | FCN3, HAKA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ficolin-3 precursor (Collagen/fibrinogen domain-containing protein 3) (Collagen/fibrinogen domain-containing lectin 3 p35) (Hakata antigen). | |||||
|
ANGP1_MOUSE
|
||||||
| NC score | 0.942851 (rank : 25) | θ value | 1.97229e-55 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O08538 | Gene names | Angpt1, Agpt | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
ANGP1_HUMAN
|
||||||
| NC score | 0.942277 (rank : 26) | θ value | 7.49467e-55 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15389 | Gene names | ANGPT1, KIAA0003 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
FCN2_HUMAN
|
||||||
| NC score | 0.929025 (rank : 27) | θ value | 3.60269e-49 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15485, Q9UC57 | Gene names | FCN2 | |||
|
Domain Architecture |

|
|||||
| Description | Ficolin-2 precursor (Collagen/fibrinogen domain-containing protein 2) (Ficolin-B) (Ficolin B) (Serum lectin p35) (EBP-37) (Hucolin) (L- Ficolin). | |||||
|
ANGL3_HUMAN
|
||||||
| NC score | 0.916821 (rank : 28) | θ value | 3.61944e-33 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y5C1 | Gene names | ANGPTL3, ANGPT5 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 3 precursor (Angiopoietin-like 3) (Angiopoietin-5) (ANG-5). | |||||
|
ANGP2_HUMAN
|
||||||
| NC score | 0.915215 (rank : 29) | θ value | 2.57589e-55 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15123, Q9NRR7, Q9P2Y7 | Gene names | ANGPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
ANGL3_MOUSE
|
||||||
| NC score | 0.911353 (rank : 30) | θ value | 7.29293e-34 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9R182 | Gene names | Angptl3 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 3 precursor (Angiopoietin-like 3). | |||||
|
ANGP2_MOUSE
|
||||||
| NC score | 0.911166 (rank : 31) | θ value | 3.36421e-55 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
FCN1_HUMAN
|
||||||
| NC score | 0.903039 (rank : 32) | θ value | 1.72781e-51 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00602, Q92596 | Gene names | FCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Ficolin-1 precursor (Collagen/fibrinogen domain-containing protein 1) (Ficolin-A) (Ficolin A) (M-Ficolin). | |||||
|
FCN1_MOUSE
|
||||||
| NC score | 0.879141 (rank : 33) | θ value | 3.15612e-45 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O70165 | Gene names | Fcn1, Fcna | |||
|
Domain Architecture |

|
|||||
| Description | Ficolin-1 precursor (Collagen/fibrinogen domain-containing protein 1) (Ficolin-A) (Ficolin A) (M-Ficolin). | |||||
|
TENN_HUMAN
|
||||||
| NC score | 0.710738 (rank : 34) | θ value | 2.95404e-43 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQP3, Q5R360 | Gene names | TNN | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
TENN_MOUSE
|
||||||
| NC score | 0.680228 (rank : 35) | θ value | 3.26607e-42 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80Z71 | Gene names | Tnn | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
TENR_HUMAN
|
||||||
| NC score | 0.668280 (rank : 36) | θ value | 4.86918e-46 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
TENR_MOUSE
|
||||||
| NC score | 0.666020 (rank : 37) | θ value | 3.15612e-45 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
TENA_MOUSE
|
||||||
| NC score | 0.554144 (rank : 38) | θ value | 1.62093e-41 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.541489 (rank : 39) | θ value | 9.50281e-42 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
TENA_HUMAN
|
||||||
| NC score | 0.540378 (rank : 40) | θ value | 1.62093e-41 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
FINC_HUMAN
|
||||||
| NC score | 0.171966 (rank : 41) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.167585 (rank : 42) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
ITL1B_MOUSE
|
||||||
| NC score | 0.153531 (rank : 43) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80ZA0 | Gene names | Itln1b, Itln2, Itlnb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intelectin-1b precursor (Intelectin-2). | |||||
|
ITL1A_MOUSE
|
||||||
| NC score | 0.150229 (rank : 44) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88310 | Gene names | Itln1a, Intl, Itln, Itlna | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intelectin-1a precursor (Intestinal lactoferrin receptor) (Galactofuranose-binding lectin). | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.142252 (rank : 45) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
COCA1_MOUSE
|
||||||
| NC score | 0.127272 (rank : 46) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
ITLN2_HUMAN
|
||||||
| NC score | 0.125173 (rank : 47) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WWU7 | Gene names | ITLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intelectin-2 precursor (Endothelial lectin HL-2). | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.124888 (rank : 48) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COEA1_HUMAN
|
||||||
| NC score | 0.121036 (rank : 49) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.119532 (rank : 50) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
COEA1_MOUSE
|
||||||
| NC score | 0.117800 (rank : 51) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
NAGPA_HUMAN
|
||||||
| NC score | 0.113543 (rank : 52) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UK23, Q96EJ8 | Gene names | NAGPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NAGPA_MOUSE
|
||||||
| NC score | 0.112538 (rank : 53) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.110635 (rank : 54) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
WIF1_HUMAN
|
||||||
| NC score | 0.105461 (rank : 55) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.105390 (rank : 56) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
WIF1_MOUSE
|
||||||
| NC score | 0.103538 (rank : 57) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.103393 (rank : 58) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.102747 (rank : 59) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
PTPRB_HUMAN
|
||||||
| NC score | 0.101130 (rank : 60) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.098422 (rank : 61) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.096944 (rank : 62) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.096902 (rank : 63) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.095325 (rank : 64) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
ITLN1_HUMAN
|
||||||
| NC score | 0.095039 (rank : 65) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WWA0, Q6YDJ3, Q9NP67 | Gene names | ITLN1, INTL, ITLN, LFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intelectin-1 precursor (Intestinal lactoferrin receptor) (Galactofuranose-binding lectin) (Endothelial lectin HL-1) (Omentin). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.090820 (rank : 66) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.088675 (rank : 67) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.086566 (rank : 68) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA44_HUMAN
|
||||||
| NC score | 0.086049 (rank : 69) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KRA11_HUMAN
|
||||||
| NC score | 0.079888 (rank : 70) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
KRA45_HUMAN
|
||||||
| NC score | 0.079422 (rank : 71) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
ITB8_HUMAN
|
||||||
| NC score | 0.077349 (rank : 72) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
ITB3_MOUSE
|
||||||
| NC score | 0.075938 (rank : 73) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54890 | Gene names | Itgb3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
ITB3_HUMAN
|
||||||
| NC score | 0.075650 (rank : 74) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05106, O15495, Q13413, Q14648, Q16499 | Gene names | ITGB3, GP3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
ITB5_MOUSE
|
||||||
| NC score | 0.075611 (rank : 75) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.075276 (rank : 76) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
ITB2_HUMAN
|
||||||
| NC score | 0.074048 (rank : 77) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05107, Q16418 | Gene names | ITGB2, CD18 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/p150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
ITB2_MOUSE
|
||||||
| NC score | 0.073309 (rank : 78) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
ITB5_HUMAN
|
||||||
| NC score | 0.073019 (rank : 79) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18084 | Gene names | ITGB5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.072850 (rank : 80) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
ITB6_HUMAN
|
||||||
| NC score | 0.072747 (rank : 81) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18564, Q16500 | Gene names | ITGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.072594 (rank : 82) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
FNDC3_MOUSE
|
||||||
| NC score | 0.071810 (rank : 83) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX90, Q2VEY7, Q6ZQ15, Q811D3, Q8BTM3 | Gene names | Fndc3a, D14Ertd453e, Fndc3, Kiaa0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.071770 (rank : 84) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
ITB6_MOUSE
|
||||||
| NC score | 0.070964 (rank : 85) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ITB7_HUMAN
|
||||||
| NC score | 0.070715 (rank : 86) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26010 | Gene names | ITGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-7 precursor. | |||||
|
ITB1_MOUSE
|
||||||
| NC score | 0.070132 (rank : 87) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
KR412_HUMAN
|
||||||
| NC score | 0.069247 (rank : 88) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
ITB1_HUMAN
|
||||||
| NC score | 0.069123 (rank : 89) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB4_HUMAN
|
||||||
| NC score | 0.069096 (rank : 90) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
KRA13_HUMAN
|
||||||
| NC score | 0.068607 (rank : 91) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KRA92_HUMAN
|
||||||
| NC score | 0.068381 (rank : 92) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.068190 (rank : 93) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
HHIP_MOUSE
|
||||||
| NC score | 0.067553 (rank : 94) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TN16, Q8C0B0, Q9WU59 | Gene names | Hhip, Hip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
ITB7_MOUSE
|
||||||
| NC score | 0.067355 (rank : 95) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26011, Q3U2M1, Q64656 | Gene names | Itgb7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-7 precursor (Integrin beta-P) (M290 IEL antigen). | |||||
|
HHIP_HUMAN
|
||||||
| NC score | 0.067187 (rank : 96) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QV1, Q6PK09, Q8NCI7, Q9BXK3, Q9H1J4, Q9H7E7 | Gene names | HHIP, HIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.067147 (rank : 97) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.066501 (rank : 98) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.065751 (rank : 99) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
EGFL9_HUMAN
|
||||||
| NC score | 0.065568 (rank : 100) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
DLK_HUMAN
|
||||||
| NC score | 0.064031 (rank : 101) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
EGFL9_MOUSE
|
||||||
| NC score | 0.063553 (rank : 102) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.061939 (rank : 103) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KRA42_HUMAN
|
||||||
| NC score | 0.061932 (rank : 104) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
DLK_MOUSE
|
||||||
| NC score | 0.061917 (rank : 105) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
RELN_MOUSE
|
||||||
| NC score | 0.061188 (rank : 106) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60841, Q9CUA6 | Gene names | Reln, Rl | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-) (Reeler protein). | |||||
|
RELN_HUMAN
|
||||||
| NC score | 0.060902 (rank : 107) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.060832 (rank : 108) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.060136 (rank : 109) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KRA99_HUMAN
|
||||||
| NC score | 0.059533 (rank : 110) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.059371 (rank : 111) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
FNDC3_HUMAN
|
||||||
| NC score | 0.058960 (rank : 112) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2H6, Q5JVF9, Q6EVH3, Q6N020, Q6P9D5, Q9H1W1 | Gene names | FNDC3A, FNDC3, KIAA0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.058875 (rank : 113) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.058531 (rank : 114) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.058406 (rank : 115) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.058130 (rank : 116) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
DNER_MOUSE
|
||||||
| NC score | 0.057907 (rank : 117) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZM4, Q3U697, Q8R226, Q8R4T6, Q8VD97 | Gene names | Dner, Bet, Bret | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor (Brain EGF repeat-containing transmembrane protein). | |||||
|
PTPRJ_HUMAN
|
||||||
| NC score | 0.057868 (rank : 118) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
DNER_HUMAN
|
||||||
| NC score | 0.057580 (rank : 119) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.056611 (rank : 120) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
DLL3_MOUSE
|
||||||
| NC score | 0.056275 (rank : 121) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.056022 (rank : 122) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
DLL4_HUMAN
|
||||||
| NC score | 0.055968 (rank : 123) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.055563 (rank : 124) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.054687 (rank : 125) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
KR414_HUMAN
|
||||||
| NC score | 0.054444 (rank : 126) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.054342 (rank : 127) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.054107 (rank : 128) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.054031 (rank : 129) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.053505 (rank : 130) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
PTPRJ_MOUSE
|
||||||
| NC score | 0.053293 (rank : 131) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.053141 (rank : 132) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
DLL1_MOUSE
|
||||||
| NC score | 0.052790 (rank : 133) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.052749 (rank : 134) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.052267 (rank : 135) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
USH2A_MOUSE
|
||||||
| NC score | 0.050206 (rank : 136) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
BCAS2_HUMAN
|
||||||
| NC score | 0.044931 (rank : 137) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75934, Q6FGS0 | Gene names | BCAS2, DAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 (DNA amplified in mammary carcinoma 1 protein) (Spliceosome-associated protein SPF 27). | |||||
|
BCAS2_MOUSE
|
||||||
| NC score | 0.044931 (rank : 138) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D287, Q3TKJ5, Q91YX8, Q9D2U4, Q9DAI0 | Gene names | Bcas2, Dam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 homolog (DNA amplified in mammary carcinoma 1 protein). | |||||
|
KCNN1_HUMAN
|
||||||
| NC score | 0.036027 (rank : 139) | θ value | 0.0563607 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| NC score | 0.033426 (rank : 140) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN2_HUMAN
|
||||||
| NC score | 0.032076 (rank : 141) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.031945 (rank : 142) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.031085 (rank : 143) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.029715 (rank : 144) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.027307 (rank : 145) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.026474 (rank : 146) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.022859 (rank : 147) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.020317 (rank : 148) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
T3JAM_MOUSE
|
||||||
| NC score | 0.020120 (rank : 149) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.019858 (rank : 150) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.019270 (rank : 151) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.019153 (rank : 152) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.017690 (rank : 153) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.017103 (rank : 154) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.016986 (rank : 155) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.016383 (rank : 156) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.016317 (rank : 157) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.016185 (rank : 158) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.016111 (rank : 159) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.015376 (rank : 160) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.014776 (rank : 161) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.014627 (rank : 162) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.012159 (rank : 163) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ST18_HUMAN
|
||||||
| NC score | 0.011131 (rank : 164) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
IDE_HUMAN
|
||||||
| NC score | 0.010881 (rank : 165) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.009883 (rank : 166) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
HUS1_MOUSE
|
||||||
| NC score | 0.008623 (rank : 167) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BQY8, O70543, Q6P8H5 | Gene names | Hus1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Checkpoint protein HUS1 (mHUS1). | |||||
|
LMNA_HUMAN
|
||||||
| NC score | 0.007104 (rank : 168) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 653 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02545, P02546, Q969I8, Q96JA2 | Gene names | LMNA, LMN1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C (70 kDa lamin) (NY-REN-32 antigen). | |||||
|
LMNA_MOUSE
|
||||||
| NC score | 0.006370 (rank : 169) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48678, P11516, P97859, Q3TIH0, Q3TTS8, Q3UCA0, Q3UCJ8, Q91WF2, Q9DC21 | Gene names | Lmna, Lmn1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C. | |||||
|
HDAC5_HUMAN
|
||||||
| NC score | 0.005391 (rank : 170) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
PLCE_MOUSE
|
||||||
| NC score | 0.004954 (rank : 171) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D1E8, Q3U702, Q8BG61, Q8CGN6 | Gene names | Agpat5, D8Ertd319e | |||
|
Domain Architecture |

|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase epsilon (EC 2.3.1.51) (1-AGP acyltransferase 5) (1-AGPAT 5) (Lysophosphatidic acid acyltransferase-epsilon) (LPAAT-epsilon) (1-acylglycerol-3-phosphate O-acyltransferase 5). | |||||
|
PLXD1_HUMAN
|
||||||
| NC score | 0.004257 (rank : 172) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
CHGUT_HUMAN
|
||||||
| NC score | 0.003863 (rank : 173) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||