Please be patient as the page loads
|
TRPM3_HUMAN
|
||||||
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TRPM3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPM6_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.960226 (rank : 4) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM6_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.959760 (rank : 5) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.961433 (rank : 3) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.961689 (rank : 2) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM2_MOUSE
|
||||||
| θ value | 1.42914e-130 (rank : 6) | NC score | 0.872066 (rank : 6) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM2_HUMAN
|
||||||
| θ value | 9.035e-101 (rank : 7) | NC score | 0.862323 (rank : 7) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM8_HUMAN
|
||||||
| θ value | 9.07703e-85 (rank : 8) | NC score | 0.855440 (rank : 8) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM8_MOUSE
|
||||||
| θ value | 1.60224e-81 (rank : 9) | NC score | 0.849207 (rank : 9) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPC4_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 10) | NC score | 0.418986 (rank : 10) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 11) | NC score | 0.417927 (rank : 11) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC5_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 12) | NC score | 0.401318 (rank : 12) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC5_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 13) | NC score | 0.400876 (rank : 13) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPV6_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 14) | NC score | 0.208121 (rank : 27) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV6_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 15) | NC score | 0.190226 (rank : 30) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 16) | NC score | 0.191439 (rank : 29) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPC2_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 17) | NC score | 0.363806 (rank : 14) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPC3_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 18) | NC score | 0.325662 (rank : 15) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPV5_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 19) | NC score | 0.180922 (rank : 33) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPC3_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 20) | NC score | 0.315065 (rank : 16) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
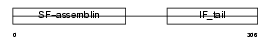
LMNA_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 21) | NC score | 0.056543 (rank : 44) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P48678, P11516, P97859, Q3TIH0, Q3TTS8, Q3UCA0, Q3UCJ8, Q91WF2, Q9DC21 | Gene names | Lmna, Lmn1 | |||
|
Domain Architecture |
|
|||||
| Description | Lamin-A/C. | |||||
|
LMNA_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 22) | NC score | 0.056737 (rank : 43) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 653 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P02545, P02546, Q969I8, Q96JA2 | Gene names | LMNA, LMN1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C (70 kDa lamin) (NY-REN-32 antigen). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 23) | NC score | 0.256566 (rank : 23) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 24) | NC score | 0.253255 (rank : 24) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 25) | NC score | 0.047659 (rank : 47) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
DREB_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 26) | NC score | 0.056458 (rank : 45) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
TRPV1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 27) | NC score | 0.185880 (rank : 31) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
LMNB2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 28) | NC score | 0.045844 (rank : 48) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
NIN_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 29) | NC score | 0.037687 (rank : 54) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
TRPV1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 30) | NC score | 0.191687 (rank : 28) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPC6_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 31) | NC score | 0.288603 (rank : 18) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 32) | NC score | 0.289685 (rank : 17) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
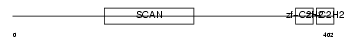
ZFP38_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 33) | NC score | 0.002257 (rank : 122) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07231 | Gene names | Znf38, Zfp-38, Zfp38, Zipro1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 38 (Zfp-38) (CtFIN51) (Transcription factor RU49). | |||||
|
DREB_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 34) | NC score | 0.050119 (rank : 46) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.029041 (rank : 61) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LMNB1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.042647 (rank : 51) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
LMNB1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 37) | NC score | 0.042857 (rank : 50) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
NIN_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 38) | NC score | 0.036518 (rank : 55) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 39) | NC score | 0.184100 (rank : 32) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 40) | NC score | 0.171642 (rank : 34) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
ZN509_HUMAN
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.001676 (rank : 125) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
CO4B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.016361 (rank : 91) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01029, Q31201, Q61372, Q61859, Q62353 | Gene names | C4b, C4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C4-B precursor [Contains: Complement C4 beta chain; Complement C4 alpha chain; C4a anaphylatoxin; Complement C4 gamma chain]. | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.038058 (rank : 53) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.032309 (rank : 57) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.018741 (rank : 81) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
CNDP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.030612 (rank : 58) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KN2, Q2TBG0, Q6UWK2, Q9BT98 | Gene names | CNDP1, CN1, CPGL2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-Ala-His dipeptidase precursor (EC 3.4.13.20) (Carnosine dipeptidase 1) (CNDP dipeptidase 1) (Serum carnosinase) (Glutamate carboxypeptidase-like protein 2). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.003740 (rank : 121) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.019111 (rank : 78) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SSNA1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.045062 (rank : 49) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ94 | Gene names | Ssna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sjoegren syndrome nuclear autoantigen 1 homolog. | |||||
|
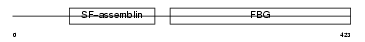
FGL2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.010758 (rank : 105) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12804 | Gene names | Fgl2, Fiblp | |||
|
Domain Architecture |
|
|||||
| Description | Fibroleukin precursor (Fibrinogen-like protein 2) (Prothrombinase) (Cytotoxic T-lymphocyte-specific protein). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.015398 (rank : 93) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
CD2L5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | -0.000665 (rank : 129) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.016682 (rank : 88) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.018933 (rank : 80) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.025486 (rank : 63) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.009722 (rank : 107) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.024347 (rank : 64) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
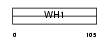
HOME1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.029672 (rank : 60) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |
|
|||||
| Description | Homer protein homolog 1. | |||||
|
TPM2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.023790 (rank : 65) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.268697 (rank : 22) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.268812 (rank : 21) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.154202 (rank : 36) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.013802 (rank : 97) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
HOME1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.030047 (rank : 59) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.016236 (rank : 92) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.014038 (rank : 96) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
TPM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.023449 (rank : 67) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.016598 (rank : 89) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
BRMS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.023239 (rank : 68) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCU9 | Gene names | BRMS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer metastasis-suppressor 1. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.022895 (rank : 69) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
PACN2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.009179 (rank : 109) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVE8 | Gene names | Pacsin2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 2. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.013088 (rank : 100) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SSNA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.036037 (rank : 56) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43805, Q6FG70, Q9BVW8 | Gene names | SSNA1, NA14 | |||
|
Domain Architecture |
|
|||||
| Description | Sjoegren syndrome nuclear autoantigen 1 (Nuclear autoantigen of 14 kDa). | |||||
|
AKAP6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.023698 (rank : 66) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
RBM13_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.021239 (rank : 74) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGS0, Q921E0, Q9D5G6 | Gene names | Rbm13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAK16-like protein RBM13 (RNA-binding motif protein 13). | |||||
|
CCD82_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.015263 (rank : 94) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PG04, Q3V462 | Gene names | Ccdc82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.008998 (rank : 110) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.027721 (rank : 62) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.020920 (rank : 75) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.002014 (rank : 123) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.012474 (rank : 102) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.001684 (rank : 124) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
ZN694_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.000064 (rank : 126) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q63HK3, Q6ZN77 | Gene names | ZNF694 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 694. | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.022845 (rank : 70) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.011752 (rank : 103) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
K1C14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.018367 (rank : 83) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
NGAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.013338 (rank : 99) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.006413 (rank : 118) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
OSTP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.015066 (rank : 95) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10923, P19008 | Gene names | Spp1, Eta-1, Op, Spp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Minopontin) (Early T-lymphocyte activation 1 protein) (2AR) (Calcium oxalate crystal growth inhibitor protein). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.017968 (rank : 86) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
TTRAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.013693 (rank : 98) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JJX7 | Gene names | Ttrap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF and TNF receptor-associated protein. | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.018343 (rank : 84) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.007924 (rank : 113) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.012802 (rank : 101) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
LMNB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.038828 (rank : 52) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P21619, P48680, Q8CGB1 | Gene names | Lmnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
LRP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.000027 (rank : 127) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.017551 (rank : 87) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.091896 (rank : 40) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PTC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.008971 (rank : 111) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35595, Q546T0 | Gene names | Ptch2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
SET_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.010884 (rank : 104) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01105, Q15541, Q5VXV1 | Gene names | SET | |||
|
Domain Architecture |
|
|||||
| Description | Protein SET (Phosphatase 2A inhibitor I2PP2A) (I-2PP2A) (Template- activating factor I) (TAF-I) (HLA-DR-associated protein II) (PHAPII) (Inhibitor of granzyme A-activated DNase) (IGAAD). | |||||
|
SET_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.010383 (rank : 106) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQU5, Q9CY82, Q9D0A9, Q9Z181 | Gene names | Set | |||
|
Domain Architecture |
|
|||||
| Description | Protein SET (Phosphatase 2A inhibitor I2PP2A) (I-2PP2A) (Template- activating factor I) (TAF-I). | |||||
|
STX11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.006504 (rank : 117) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75558, O75378, O95148 | Gene names | STX11 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-11. | |||||
|
TRPC7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.277302 (rank : 20) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.278300 (rank : 19) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |
|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.006670 (rank : 116) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
ZXDA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | -0.000455 (rank : 128) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98168, Q9UJP7 | Gene names | ZXDA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger X-linked protein ZXDA. | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.007312 (rank : 115) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
FMN1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.006335 (rank : 119) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
FMN1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.005603 (rank : 120) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.009336 (rank : 108) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.022186 (rank : 71) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.021638 (rank : 72) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LIPB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.019312 (rank : 77) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.020136 (rank : 76) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
PTC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.008132 (rank : 112) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6C5, O95341, O95856, Q5QP87, Q6UX14 | Gene names | PTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.021251 (rank : 73) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RBM13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.016371 (rank : 90) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXY0, Q5U5T1, Q86UC4, Q96SY6 | Gene names | RBM13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAK16-like protein RBM13 (RNA-binding motif protein 13) (NNP78). | |||||
|
REST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.018692 (rank : 82) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.018053 (rank : 85) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.007616 (rank : 114) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.019086 (rank : 79) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
EF2K_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.208167 (rank : 26) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EF2K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.216092 (rank : 25) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
NUD9P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.115862 (rank : 39) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N2Z3 | Gene names | NUDT9P1, C10orf98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative nucleoside diphosphate-linked moiety X motif 9P1. | |||||
|
NUDT9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.153386 (rank : 37) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BW91, Q8NBN1, Q8NCB9, Q8NG25 | Gene names | NUDT9, NUDT10 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
NUDT9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.154252 (rank : 35) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.058195 (rank : 42) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.061651 (rank : 41) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
TRPV2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.129955 (rank : 38) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
TRPM3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.961689 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.961433 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM6_HUMAN
|
||||||
| NC score | 0.960226 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM6_MOUSE
|
||||||
| NC score | 0.959760 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM2_MOUSE
|
||||||
| NC score | 0.872066 (rank : 6) | θ value | 1.42914e-130 (rank : 6) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM2_HUMAN
|
||||||
| NC score | 0.862323 (rank : 7) | θ value | 9.035e-101 (rank : 7) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM8_HUMAN
|
||||||
| NC score | 0.855440 (rank : 8) | θ value | 9.07703e-85 (rank : 8) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM8_MOUSE
|
||||||
| NC score | 0.849207 (rank : 9) | θ value | 1.60224e-81 (rank : 9) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.418986 (rank : 10) | θ value | 5.43371e-13 (rank : 10) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.417927 (rank : 11) | θ value | 7.09661e-13 (rank : 11) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC5_MOUSE
|
||||||
| NC score | 0.401318 (rank : 12) | θ value | 2.79066e-09 (rank : 12) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC5_HUMAN
|
||||||
| NC score | 0.400876 (rank : 13) | θ value | 1.06045e-08 (rank : 13) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC2_MOUSE
|
||||||
| NC score | 0.363806 (rank : 14) | θ value | 8.40245e-06 (rank : 17) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPC3_MOUSE
|
||||||
| NC score | 0.325662 (rank : 15) | θ value | 1.43324e-05 (rank : 18) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC3_HUMAN
|
||||||
| NC score | 0.315065 (rank : 16) | θ value | 9.29e-05 (rank : 20) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.289685 (rank : 17) | θ value | 0.0431538 (rank : 32) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC6_HUMAN
|
||||||
| NC score | 0.288603 (rank : 18) | θ value | 0.0431538 (rank : 31) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC7_MOUSE
|
||||||
| NC score | 0.278300 (rank : 19) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC7_HUMAN
|
||||||
| NC score | 0.277302 (rank : 20) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.268812 (rank : 21) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.268697 (rank : 22) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.256566 (rank : 23) | θ value | 0.00134147 (rank : 23) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.253255 (rank : 24) | θ value | 0.00175202 (rank : 24) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
EF2K_MOUSE
|
||||||
| NC score | 0.216092 (rank : 25) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EF2K_HUMAN
|
||||||
| NC score | 0.208167 (rank : 26) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
TRPV6_HUMAN
|
||||||
| NC score | 0.208121 (rank : 27) | θ value | 9.92553e-07 (rank : 14) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV1_HUMAN
|
||||||
| NC score | 0.191687 (rank : 28) | θ value | 0.0252991 (rank : 30) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.191439 (rank : 29) | θ value | 4.92598e-06 (rank : 16) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV6_MOUSE
|
||||||
| NC score | 0.190226 (rank : 30) | θ value | 2.88788e-06 (rank : 15) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
TRPV1_MOUSE
|
||||||
| NC score | 0.185880 (rank : 31) | θ value | 0.00869519 (rank : 27) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.184100 (rank : 32) | θ value | 0.0961366 (rank : 39) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV5_MOUSE
|
||||||
| NC score | 0.180922 (rank : 33) | θ value | 3.19293e-05 (rank : 19) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.171642 (rank : 34) | θ value | 0.0961366 (rank : 40) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
NUDT9_MOUSE
|
||||||
| NC score | 0.154252 (rank : 35) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.154202 (rank : 36) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
NUDT9_HUMAN
|
||||||
| NC score | 0.153386 (rank : 37) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BW91, Q8NBN1, Q8NCB9, Q8NG25 | Gene names | NUDT9, NUDT10 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
TRPV2_HUMAN
|
||||||
| NC score | 0.129955 (rank : 38) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
NUD9P_HUMAN
|
||||||
| NC score | 0.115862 (rank : 39) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N2Z3 | Gene names | NUDT9P1, C10orf98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative nucleoside diphosphate-linked moiety X motif 9P1. | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.091896 (rank : 40) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.061651 (rank : 41) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.058195 (rank : 42) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
LMNA_HUMAN
|
||||||
| NC score | 0.056737 (rank : 43) | θ value | 0.000270298 (rank : 22) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 653 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P02545, P02546, Q969I8, Q96JA2 | Gene names | LMNA, LMN1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C (70 kDa lamin) (NY-REN-32 antigen). | |||||
|
LMNA_MOUSE
|
||||||
| NC score | 0.056543 (rank : 44) | θ value | 0.000158464 (rank : 21) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P48678, P11516, P97859, Q3TIH0, Q3TTS8, Q3UCA0, Q3UCJ8, Q91WF2, Q9DC21 | Gene names | Lmna, Lmn1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C. | |||||
|
DREB_HUMAN
|
||||||
| NC score | 0.056458 (rank : 45) | θ value | 0.00665767 (rank : 26) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
DREB_MOUSE
|
||||||
| NC score | 0.050119 (rank : 46) | θ value | 0.0736092 (rank : 34) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.047659 (rank : 47) | θ value | 0.00665767 (rank : 25) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
LMNB2_HUMAN
|
||||||
| NC score | 0.045844 (rank : 48) | θ value | 0.0148317 (rank : 28) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
SSNA1_MOUSE
|
||||||
| NC score | 0.045062 (rank : 49) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ94 | Gene names | Ssna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sjoegren syndrome nuclear autoantigen 1 homolog. | |||||
|
LMNB1_MOUSE
|
||||||
| NC score | 0.042857 (rank : 50) | θ value | 0.0961366 (rank : 37) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
LMNB1_HUMAN
|
||||||
| NC score | 0.042647 (rank : 51) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
LMNB2_MOUSE
|
||||||
| NC score | 0.038828 (rank : 52) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P21619, P48680, Q8CGB1 | Gene names | Lmnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.038058 (rank : 53) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.037687 (rank : 54) | θ value | 0.0252991 (rank : 29) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.036518 (rank : 55) | θ value | 0.0961366 (rank : 38) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
SSNA1_HUMAN
|
||||||
| NC score | 0.036037 (rank : 56) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43805, Q6FG70, Q9BVW8 | Gene names | SSNA1, NA14 | |||
|
Domain Architecture |

|
|||||
| Description | Sjoegren syndrome nuclear autoantigen 1 (Nuclear autoantigen of 14 kDa). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.032309 (rank : 57) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CNDP1_HUMAN
|
||||||
| NC score | 0.030612 (rank : 58) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KN2, Q2TBG0, Q6UWK2, Q9BT98 | Gene names | CNDP1, CN1, CPGL2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-Ala-His dipeptidase precursor (EC 3.4.13.20) (Carnosine dipeptidase 1) (CNDP dipeptidase 1) (Serum carnosinase) (Glutamate carboxypeptidase-like protein 2). | |||||
|
HOME1_MOUSE
|
||||||
| NC score | 0.030047 (rank : 59) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
HOME1_HUMAN
|
||||||
| NC score | 0.029672 (rank : 60) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.029041 (rank : 61) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.027721 (rank : 62) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.025486 (rank : 63) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.024347 (rank : 64) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
TPM2_MOUSE
|
||||||
| NC score | 0.023790 (rank : 65) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
AKAP6_HUMAN
|
||||||
| NC score | 0.023698 (rank : 66) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
TPM2_HUMAN
|
||||||
| NC score | 0.023449 (rank : 67) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
BRMS1_HUMAN
|
||||||
| NC score | 0.023239 (rank : 68) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCU9 | Gene names | BRMS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer metastasis-suppressor 1. | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.022895 (rank : 69) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.022845 (rank : 70) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.022186 (rank : 71) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.021638 (rank : 72) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.021251 (rank : 73) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RBM13_MOUSE
|
||||||
| NC score | 0.021239 (rank : 74) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGS0, Q921E0, Q9D5G6 | Gene names | Rbm13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAK16-like protein RBM13 (RNA-binding motif protein 13). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.020920 (rank : 75) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.020136 (rank : 76) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
LIPB2_HUMAN
|
||||||
| NC score | 0.019312 (rank : 77) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.019111 (rank : 78) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.019086 (rank : 79) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.018933 (rank : 80) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.018741 (rank : 81) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.018692 (rank : 82) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
K1C14_HUMAN
|
||||||
| NC score | 0.018367 (rank : 83) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.018343 (rank : 84) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.018053 (rank : 85) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.017968 (rank : 86) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.017551 (rank : 87) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.016682 (rank : 88) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.016598 (rank : 89) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
RBM13_HUMAN
|
||||||
| NC score | 0.016371 (rank : 90) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXY0, Q5U5T1, Q86UC4, Q96SY6 | Gene names | RBM13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAK16-like protein RBM13 (RNA-binding motif protein 13) (NNP78). | |||||
|
CO4B_MOUSE
|
||||||
| NC score | 0.016361 (rank : 91) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01029, Q31201, Q61372, Q61859, Q62353 | Gene names | C4b, C4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C4-B precursor [Contains: Complement C4 beta chain; Complement C4 alpha chain; C4a anaphylatoxin; Complement C4 gamma chain]. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.016236 (rank : 92) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.015398 (rank : 93) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
CCD82_MOUSE
|
||||||
| NC score | 0.015263 (rank : 94) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PG04, Q3V462 | Gene names | Ccdc82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
OSTP_MOUSE
|
||||||
| NC score | 0.015066 (rank : 95) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10923, P19008 | Gene names | Spp1, Eta-1, Op, Spp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Minopontin) (Early T-lymphocyte activation 1 protein) (2AR) (Calcium oxalate crystal growth inhibitor protein). | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.014038 (rank : 96) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.013802 (rank : 97) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
TTRAP_MOUSE
|
||||||
| NC score | 0.013693 (rank : 98) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JJX7 | Gene names | Ttrap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF and TNF receptor-associated protein. | |||||
|
NGAP_HUMAN
|
||||||
| NC score | 0.013338 (rank : 99) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.013088 (rank : 100) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.012802 (rank : 101) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.012474 (rank : 102) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.011752 (rank : 103) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
SET_HUMAN
|
||||||
| NC score | 0.010884 (rank : 104) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01105, Q15541, Q5VXV1 | Gene names | SET | |||
|
Domain Architecture |

|
|||||
| Description | Protein SET (Phosphatase 2A inhibitor I2PP2A) (I-2PP2A) (Template- activating factor I) (TAF-I) (HLA-DR-associated protein II) (PHAPII) (Inhibitor of granzyme A-activated DNase) (IGAAD). | |||||
|
FGL2_MOUSE
|
||||||
| NC score | 0.010758 (rank : 105) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12804 | Gene names | Fgl2, Fiblp | |||
|
Domain Architecture |

|
|||||
| Description | Fibroleukin precursor (Fibrinogen-like protein 2) (Prothrombinase) (Cytotoxic T-lymphocyte-specific protein). | |||||
|
SET_MOUSE
|
||||||
| NC score | 0.010383 (rank : 106) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQU5, Q9CY82, Q9D0A9, Q9Z181 | Gene names | Set | |||
|
Domain Architecture |

|
|||||
| Description | Protein SET (Phosphatase 2A inhibitor I2PP2A) (I-2PP2A) (Template- activating factor I) (TAF-I). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.009722 (rank : 107) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.009336 (rank : 108) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
PACN2_MOUSE
|
||||||
| NC score | 0.009179 (rank : 109) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVE8 | Gene names | Pacsin2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 2. | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.008998 (rank : 110) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
PTC2_MOUSE
|
||||||
| NC score | 0.008971 (rank : 111) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35595, Q546T0 | Gene names | Ptch2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
PTC2_HUMAN
|
||||||
| NC score | 0.008132 (rank : 112) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6C5, O95341, O95856, Q5QP87, Q6UX14 | Gene names | PTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.007924 (rank : 113) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.007616 (rank : 114) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.007312 (rank : 115) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.006670 (rank : 116) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
STX11_HUMAN
|
||||||
| NC score | 0.006504 (rank : 117) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75558, O75378, O95148 | Gene names | STX11 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-11. | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.006413 (rank : 118) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
FMN1A_MOUSE
|
||||||
| NC score | 0.006335 (rank : 119) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
FMN1B_MOUSE
|
||||||
| NC score | 0.005603 (rank : 120) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.003740 (rank : 121) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
ZFP38_MOUSE
|
||||||
| NC score | 0.002257 (rank : 122) | θ value | 0.0431538 (rank : 33) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07231 | Gene names | Znf38, Zfp-38, Zfp38, Zipro1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 38 (Zfp-38) (CtFIN51) (Transcription factor RU49). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.002014 (rank : 123) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | 0.001684 (rank : 124) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
ZN509_HUMAN
|
||||||
| NC score | 0.001676 (rank : 125) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN694_HUMAN
|
||||||
| NC score | 0.000064 (rank : 126) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q63HK3, Q6ZN77 | Gene names | ZNF694 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 694. | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.000027 (rank : 127) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
ZXDA_HUMAN
|
||||||
| NC score | -0.000455 (rank : 128) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98168, Q9UJP7 | Gene names | ZXDA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger X-linked protein ZXDA. | |||||
|
CD2L5_MOUSE
|
||||||
| NC score | -0.000665 (rank : 129) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||