Please be patient as the page loads
|
TRPC4_HUMAN
|
||||||
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TRPC4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999606 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 133 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.990147 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC5_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.990570 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | 5.1754e-181 (rank : 5) | NC score | 0.959276 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | 1.96665e-180 (rank : 6) | NC score | 0.959458 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC3_HUMAN
|
||||||
| θ value | 1.85788e-146 (rank : 7) | NC score | 0.947853 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC3_MOUSE
|
||||||
| θ value | 5.40562e-146 (rank : 8) | NC score | 0.949441 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC6_HUMAN
|
||||||
| θ value | 1.62758e-142 (rank : 9) | NC score | 0.940328 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC7_HUMAN
|
||||||
| θ value | 4.00889e-141 (rank : 10) | NC score | 0.939770 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC7_MOUSE
|
||||||
| θ value | 6.83811e-141 (rank : 11) | NC score | 0.940575 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |
|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 1.1664e-140 (rank : 12) | NC score | 0.942084 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC2_MOUSE
|
||||||
| θ value | 1.53757e-108 (rank : 13) | NC score | 0.948122 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPM8_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 14) | NC score | 0.590135 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM8_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 15) | NC score | 0.592624 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 16) | NC score | 0.496503 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 17) | NC score | 0.494015 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM6_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 18) | NC score | 0.487317 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM6_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 19) | NC score | 0.464440 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM2_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 20) | NC score | 0.503932 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM2_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 21) | NC score | 0.479442 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM3_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 22) | NC score | 0.418986 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 23) | NC score | 0.320433 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 24) | NC score | 0.339361 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPV1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 25) | NC score | 0.289959 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPV1_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 26) | NC score | 0.287413 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV5_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 27) | NC score | 0.249350 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 28) | NC score | 0.238027 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 29) | NC score | 0.195181 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 30) | NC score | 0.176229 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
TRPV2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 31) | NC score | 0.191411 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 32) | NC score | 0.194228 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
TRPV6_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 33) | NC score | 0.242584 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 34) | NC score | 0.193761 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
TRPV6_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 35) | NC score | 0.232660 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 36) | NC score | 0.207212 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 37) | NC score | 0.215779 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 38) | NC score | 0.055443 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 39) | NC score | 0.051947 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 40) | NC score | 0.241309 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 41) | NC score | 0.080270 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
AN36B_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 42) | NC score | 0.097821 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8N2N9, Q6IPR0 | Gene names | ANKRD36B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 36B. | |||||
|
RN5A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 43) | NC score | 0.093978 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q05823 | Gene names | RNASEL, RNS4 | |||
|
Domain Architecture |

|
|||||
| Description | 2-5A-dependent ribonuclease (EC 3.1.26.-) (2-5A-dependent RNase) (Ribonuclease L) (RNase L) (Ribonuclease 4). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 44) | NC score | 0.081046 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 45) | NC score | 0.223589 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
ASB15_HUMAN
|
||||||
| θ value | 0.125558 (rank : 46) | NC score | 0.067968 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8WXK1 | Gene names | ASB15 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
PCDGC_HUMAN
|
||||||
| θ value | 0.163984 (rank : 47) | NC score | 0.009566 (rank : 181) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60330, O15100, Q6UW70, Q9Y5D7 | Gene names | PCDHGA12, CDH21, FIB3, KIAA0588 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A12 precursor (PCDH-gamma-A12) (Cadherin-21) (Fibroblast cadherin 3). | |||||
|
ASZ1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 48) | NC score | 0.079705 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8VD46, Q9JKQ7 | Gene names | Asz1, Gasz | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat, SAM and basic leucine zipper domain-containing protein 1 (Germ cell-specific ankyrin, SAM and basic leucine zipper domain- containing protein). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 49) | NC score | 0.092753 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
RN5A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 50) | NC score | 0.088950 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q05921, Q9ERU7 | Gene names | Rnasel, Rns4 | |||
|
Domain Architecture |

|
|||||
| Description | 2-5A-dependent ribonuclease (EC 3.1.26.-) (2-5A-dependent RNase) (Ribonuclease L) (RNase L) (Ribonuclease 4). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.046856 (rank : 156) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
ANKY2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 52) | NC score | 0.115659 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
POTE8_HUMAN
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | 0.087261 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q6S8J7, Q6S8J6 | Gene names | POTE8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 8. | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.084214 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ANKK1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.041254 (rank : 164) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1075 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8NFD2 | Gene names | ANKK1, PKK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1) (Protein kinase PKK2) (X-kinase) (SgK288). | |||||
|
ANR18_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.053983 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
ASZ1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.072848 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WWH4 | Gene names | ASZ1, GASZ | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat, SAM and basic leucine zipper domain-containing protein 1 (Germ cell-specific ankyrin, SAM and basic leucine zipper domain- containing protein). | |||||
|
ANK1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.110635 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.110416 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.086574 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
MPP10_MOUSE
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.031948 (rank : 169) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
PCDG7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.007995 (rank : 184) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5G6, Q9Y5D0 | Gene names | PCDHGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A7 precursor (PCDH-gamma-A7). | |||||
|
ACBD6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.110432 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9D061, Q3TMN7, Q9DCU4 | Gene names | Acbd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.088208 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.065062 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.049368 (rank : 155) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
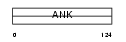
ANKR7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.076149 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q92527, Q96QN1, Q9UDM3 | Gene names | ANKRD7 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 7 (Testis-specific protein TSA806). | |||||
|
ANR19_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.063050 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H560 | Gene names | ANKRD19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 19. | |||||
|
ANR44_HUMAN
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.082537 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ANR52_HUMAN
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.091620 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| θ value | 0.813845 (rank : 71) | NC score | 0.091430 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.064793 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
PCDGB_HUMAN
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.008936 (rank : 182) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5H2, Q9Y5D8, Q9Y5D9 | Gene names | PCDHGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A11 precursor (PCDH-gamma-A11). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 0.813845 (rank : 74) | NC score | 0.077974 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
TR143_MOUSE
|
||||||
| θ value | 0.813845 (rank : 75) | NC score | 0.026103 (rank : 171) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQB9, Q2NL43 | Gene names | Tas2r143, T2r36, Tas2r43 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 143 (T2R143) (Taste receptor type 2 member 43) (T2R43) (mT2R36). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.074263 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANR21_HUMAN
|
||||||
| θ value | 1.06291 (rank : 77) | NC score | 0.079884 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
POT14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 78) | NC score | 0.069849 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.051671 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.004000 (rank : 189) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
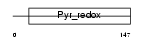
CT026_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.043068 (rank : 159) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NHU2, Q5TE18, Q8N5R9, Q96M59, Q9BQL2, Q9H127, Q9H128, Q9NQH4, Q9UFV8, Q9Y4V7 | Gene names | C20orf26 | |||
|
Domain Architecture |
|
|||||
| Description | Uncharacterized protein C20orf26. | |||||
|
NET4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.007054 (rank : 185) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HB63, Q658K9, Q7L3F1, Q7L9D6, Q7Z5B6, Q9BZP1, Q9NT44, Q9P133 | Gene names | NTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin) (Hepar-derived netrin-like protein). | |||||
|
NFKB2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.053746 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.041901 (rank : 162) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.043550 (rank : 158) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
ANR28_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.069572 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ASB15_MOUSE
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.059516 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8VHS6 | Gene names | Asb15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
MIB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.067045 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
MIB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.067151 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
AN13A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.068589 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8IZ07, O60736 | Gene names | ANKRD13A, ANKRD13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13A (Protein KE03). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.051687 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
ANKY2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.092289 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8IV38, Q96BL3 | Gene names | ANKMY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.042916 (rank : 160) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.001012 (rank : 195) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.017735 (rank : 175) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
ANKK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.036062 (rank : 167) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8BZ25 | Gene names | Ankk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1). | |||||
|
ANR28_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.068193 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANFY1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.074033 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANR15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.069587 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q14678, Q5W0W0, Q8IY65, Q8WX74 | Gene names | ANKRD15, KANK, KIAA0172 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 15 (Kidney ankyrin repeat- containing protein). | |||||
|
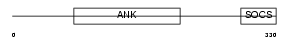
ASB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.072806 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9WV74 | Gene names | Asb1 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 1 (ASB-1). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.041955 (rank : 161) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CDC37_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.016075 (rank : 177) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
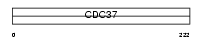
CDC37_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.018347 (rank : 174) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |
|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
RBBP8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.011347 (rank : 180) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
TRPA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.097230 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
VPS18_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.017024 (rank : 176) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R307, Q8BGV6, Q8BZX6, Q8C126 | Gene names | Vps18 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18. | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.057915 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CTNS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.019310 (rank : 173) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57757 | Gene names | Ctns | |||
|
Domain Architecture |
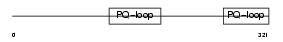
|
|||||
| Description | Cystinosin. | |||||
|
FANK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.062021 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8TC84, Q6UXY9, Q6X7T6 | Gene names | FANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type 3 and ankyrin repeat domains protein 1. | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.003713 (rank : 192) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.045170 (rank : 157) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.008395 (rank : 183) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TRPA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.105574 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.038705 (rank : 165) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
ACBD6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.086252 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BR61 | Gene names | ACBD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
AN36A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.059981 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6UX02, Q86X62 | Gene names | ANKRD36, ANKRD36A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 36A. | |||||
|
ANKS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.090567 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
ANKS6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.078460 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
ASB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.089413 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9Y575, Q9NVZ2 | Gene names | ASB3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
BCL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.064126 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |
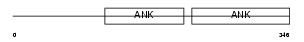
|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.034900 (rank : 168) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.036212 (rank : 166) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CLPB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.050937 (rank : 150) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H078, Q8ND11, Q9H8Y0 | Gene names | CLPB, SKD3 | |||
|
Domain Architecture |

|
|||||
| Description | Caseinolytic peptidase B protein homolog (Suppressor of potassium transport defect 3). | |||||
|
DOPO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.011835 (rank : 179) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09172, Q5T381, Q96AG2 | Gene names | DBH | |||
|
Domain Architecture |

|
|||||
| Description | Dopamine beta-hydroxylase precursor (EC 1.14.17.1) (Dopamine beta- monooxygenase). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.003836 (rank : 191) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
K0831_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.006916 (rank : 186) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
LZTS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.006802 (rank : 187) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.003992 (rank : 190) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
PSD10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.053686 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |
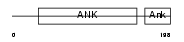
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.003557 (rank : 193) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.005238 (rank : 188) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ANKF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.019733 (rank : 172) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N957 | Gene names | ANKFN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and fibronectin type-III domain-containing protein 1. | |||||
|
ANKS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.083908 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P59672, Q6ZQG0 | Gene names | Anks1a, Anks1, Kiaa0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A. | |||||
|
ASB17_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.012271 (rank : 178) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHP9, Q9CUF9 | Gene names | Asb17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 17 (ASB-17). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.071070 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.072300 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.027609 (rank : 170) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
RIPK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.041723 (rank : 163) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1094 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P57078, Q96KH0 | Gene names | RIPK4, ANKRD3, DIK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 4 (EC 2.7.11.1) (Ankyrin repeat domain protein 3) (PKC-delta-interacting protein kinase). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | -0.003605 (rank : 196) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.002161 (rank : 194) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
ANFY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.054859 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANKR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.053486 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANKR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.068282 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANKR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.052575 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9GZV1, Q5T456, Q8WUD7 | Gene names | ANKRD2, ARPP | |||
|
Domain Architecture |
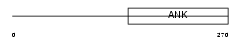
|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (hArpp). | |||||
|
ANKR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.051685 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9WV06 | Gene names | Ankrd2, Arpp | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp). | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.055061 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.072913 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
ANR16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.053067 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6P6B7, Q9NT01 | Gene names | ANKRD16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 16. | |||||
|
ANR22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.050689 (rank : 151) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
ANR25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.053147 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
ANR29_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.072690 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N6D5, Q6ZWE8, Q96LU9 | Gene names | ANKRD29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 29. | |||||
|
ANR32_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.053103 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8R3P9, Q5XK30, Q8BHY5, Q921Y9 | Gene names | Ankrd32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 32. | |||||
|
ANR35_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.065347 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
ANR46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.050958 (rank : 149) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q86W74, Q6P9B7 | Gene names | ANKRD46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
ANR49_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.051164 (rank : 147) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8VE42, Q9Z2S1 | Gene names | Ankrd49, Fgif, Gbif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor) (Fetal globin-increasing factor). | |||||
|
ANR50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.080476 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ANRA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.052912 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANRA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.053271 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ASB14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.056444 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8VHS7 | Gene names | Asb14 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 14 (ASB-14). | |||||
|
ASB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.062307 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y576, Q9ULS4 | Gene names | ASB1, KIAA1146 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 1 (ASB-1). | |||||
|
ASB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.065944 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96Q27, Q9NSU5, Q9Y567 | Gene names | ASB2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 2 (ASB-2). | |||||
|
ASB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.062339 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8K0L0, Q9CTH4, Q9WV73 | Gene names | Asb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 2 (ASB-2). | |||||
|
ASB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.080434 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.053176 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NWX5 | Gene names | ASB6 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
ASB6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.052272 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q91ZU1, Q8VEC6 | Gene names | Asb6 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
ASB7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.054285 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H672, Q7Z4S3 | Gene names | ASB7 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.054247 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q91ZU0 | Gene names | Asb7 | |||
|
Domain Architecture |
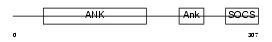
|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.050540 (rank : 152) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
Domain Architecture |
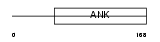
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
BARD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.065328 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
BARD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.077560 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.054462 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.073149 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.072270 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CT012_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.056796 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8C008, Q8C060 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C20orf12 homolog. | |||||
|
EF2K_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.050527 (rank : 153) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |
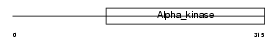
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EF2K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.052500 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |
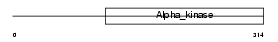
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.066860 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
FANK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.054353 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9DAM9, Q9CUA7 | Gene names | Fank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type 3 and ankyrin repeat domains 1 protein (Germ cell- specific gene 1 protein) (GSG1). | |||||
|
GABP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.068961 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |
|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
GABP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.072309 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q00420, Q00421, Q8BS31, Q91YZ0, Q9QVV2 | Gene names | Gabpb2, Gabpb, Gabpb1 | |||
|
Domain Architecture |
|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1). | |||||
|
IKBA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.052761 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P25963 | Gene names | NFKBIA, IKBA, MAD3, NFKBI | |||
|
Domain Architecture |
|
|||||
| Description | NF-kappa-B inhibitor alpha (Major histocompatibility complex enhancer- binding protein MAD3) (I-kappa-B-alpha) (IkappaBalpha) (IKB-alpha). | |||||
|
IKBA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.052643 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Z1E3, Q80ZX5 | Gene names | Nfkbia, Ikba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B inhibitor alpha (I-kappa-B-alpha) (IkappaBalpha) (IKB- alpha). | |||||
|
IKBB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.051348 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
IKBB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.051004 (rank : 148) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
IKBE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.053429 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
INVS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.061928 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
INVS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.062775 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
MIB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.053922 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
MIB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.053506 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.054300 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.059688 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
PA2G6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.054227 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PA2G6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.061136 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
POT22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.054919 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6S545 | Gene names | ACTBL1, POTE22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 22. | |||||
|
PSD10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.050305 (rank : 154) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.061145 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.999606 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 133 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC5_MOUSE
|
||||||
| NC score | 0.990570 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC5_HUMAN
|
||||||
| NC score | 0.990147 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.959458 (rank : 5) | θ value | 1.96665e-180 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.959276 (rank : 6) | θ value | 5.1754e-181 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC3_MOUSE
|
||||||
| NC score | 0.949441 (rank : 7) | θ value | 5.40562e-146 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC2_MOUSE
|
||||||
| NC score | 0.948122 (rank : 8) | θ value | 1.53757e-108 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPC3_HUMAN
|
||||||
| NC score | 0.947853 (rank : 9) | θ value | 1.85788e-146 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.942084 (rank : 10) | θ value | 1.1664e-140 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC7_MOUSE
|
||||||
| NC score | 0.940575 (rank : 11) | θ value | 6.83811e-141 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC6_HUMAN
|
||||||
| NC score | 0.940328 (rank : 12) | θ value | 1.62758e-142 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC7_HUMAN
|
||||||
| NC score | 0.939770 (rank : 13) | θ value | 4.00889e-141 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPM8_MOUSE
|
||||||
| NC score | 0.592624 (rank : 14) | θ value | 2.87452e-22 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPM8_HUMAN
|
||||||
| NC score | 0.590135 (rank : 15) | θ value | 3.39556e-23 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM2_HUMAN
|
||||||
| NC score | 0.503932 (rank : 16) | θ value | 4.91457e-14 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.496503 (rank : 17) | θ value | 6.20254e-17 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.494015 (rank : 18) | θ value | 4.02038e-16 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM6_MOUSE
|
||||||
| NC score | 0.487317 (rank : 19) | θ value | 5.25075e-16 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM2_MOUSE
|
||||||
| NC score | 0.479442 (rank : 20) | θ value | 4.91457e-14 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM6_HUMAN
|
||||||
| NC score | 0.464440 (rank : 21) | θ value | 4.44505e-15 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM3_HUMAN
|
||||||
| NC score | 0.418986 (rank : 22) | θ value | 5.43371e-13 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.339361 (rank : 23) | θ value | 4.45536e-07 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.320433 (rank : 24) | θ value | 1.53129e-07 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV1_MOUSE
|
||||||
| NC score | 0.289959 (rank : 25) | θ value | 2.21117e-06 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPV1_HUMAN
|
||||||
| NC score | 0.287413 (rank : 26) | θ value | 4.92598e-06 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV5_MOUSE
|
||||||
| NC score | 0.249350 (rank : 27) | θ value | 0.00020696 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV6_HUMAN
|
||||||
| NC score | 0.242584 (rank : 28) | θ value | 0.00298849 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.241309 (rank : 29) | θ value | 0.0431538 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.238027 (rank : 30) | θ value | 0.000270298 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
TRPV6_MOUSE
|
||||||
| NC score | 0.232660 (rank : 31) | θ value | 0.00665767 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.223589 (rank : 32) | θ value | 0.0961366 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.215779 (rank : 33) | θ value | 0.0113563 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.207212 (rank : 34) | θ value | 0.0113563 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.195181 (rank : 35) | θ value | 0.000270298 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.194228 (rank : 36) | θ value | 0.00134147 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.193761 (rank : 37) | θ value | 0.00665767 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
TRPV2_HUMAN
|
||||||
| NC score | 0.191411 (rank : 38) | θ value | 0.00102713 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.176229 (rank : 39) | θ value | 0.00102713 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
ANKY2_MOUSE
|
||||||
| NC score | 0.115659 (rank : 40) | θ value | 0.279714 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.110635 (rank : 41) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ACBD6_MOUSE
|
||||||
| NC score | 0.110432 (rank : 42) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9D061, Q3TMN7, Q9DCU4 | Gene names | Acbd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.110416 (rank : 43) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
TRPA1_HUMAN
|
||||||
| NC score | 0.105574 (rank : 44) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
AN36B_HUMAN
|
||||||
| NC score | 0.097821 (rank : 45) | θ value | 0.0736092 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8N2N9, Q6IPR0 | Gene names | ANKRD36B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 36B. | |||||
|
TRPA1_MOUSE
|
||||||
| NC score | 0.097230 (rank : 46) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
RN5A_HUMAN
|
||||||
| NC score | 0.093978 (rank : 47) | θ value | 0.0961366 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q05823 | Gene names | RNASEL, RNS4 | |||
|
Domain Architecture |

|
|||||
| Description | 2-5A-dependent ribonuclease (EC 3.1.26.-) (2-5A-dependent RNase) (Ribonuclease L) (RNase L) (Ribonuclease 4). | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.092753 (rank : 48) | θ value | 0.21417 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
ANKY2_HUMAN
|
||||||
| NC score | 0.092289 (rank : 49) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8IV38, Q96BL3 | Gene names | ANKMY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ANR52_HUMAN
|
||||||
| NC score | 0.091620 (rank : 50) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| NC score | 0.091430 (rank : 51) | θ value | 0.813845 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANKS1_HUMAN
|
||||||
| NC score | 0.090567 (rank : 52) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
ASB3_HUMAN
|
||||||
| NC score | 0.089413 (rank : 53) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9Y575, Q9NVZ2 | Gene names | ASB3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
RN5A_MOUSE
|
||||||
| NC score | 0.088950 (rank : 54) | θ value | 0.21417 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q05921, Q9ERU7 | Gene names | Rnasel, Rns4 | |||
|
Domain Architecture |

|
|||||
| Description | 2-5A-dependent ribonuclease (EC 3.1.26.-) (2-5A-dependent RNase) (Ribonuclease L) (RNase L) (Ribonuclease 4). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.088208 (rank : 55) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
POTE8_HUMAN
|
||||||
| NC score | 0.087261 (rank : 56) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q6S8J7, Q6S8J6 | Gene names | POTE8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 8. | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.086574 (rank : 57) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
ACBD6_HUMAN
|
||||||
| NC score | 0.086252 (rank : 58) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BR61 | Gene names | ACBD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.084214 (rank : 59) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ANKS1_MOUSE
|
||||||
| NC score | 0.083908 (rank : 60) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P59672, Q6ZQG0 | Gene names | Anks1a, Anks1, Kiaa0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A. | |||||
|
ANR44_HUMAN
|
||||||
| NC score | 0.082537 (rank : 61) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.081046 (rank : 62) | θ value | 0.0961366 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
ANR50_HUMAN
|
||||||
| NC score | 0.080476 (rank : 63) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ASB3_MOUSE
|
||||||
| NC score | 0.080434 (rank : 64) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.080270 (rank : 65) | θ value | 0.0563607 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
ANR21_HUMAN
|
||||||
| NC score | 0.079884 (rank : 66) | θ value | 1.06291 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
ASZ1_MOUSE
|
||||||
| NC score | 0.079705 (rank : 67) | θ value | 0.21417 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8VD46, Q9JKQ7 | Gene names | Asz1, Gasz | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat, SAM and basic leucine zipper domain-containing protein 1 (Germ cell-specific ankyrin, SAM and basic leucine zipper domain- containing protein). | |||||
|
ANKS6_HUMAN
|
||||||
| NC score | 0.078460 (rank : 68) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.077974 (rank : 69) | θ value | 0.813845 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
BARD1_MOUSE
|
||||||
| NC score | 0.077560 (rank : 70) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
ANKR7_HUMAN
|
||||||
| NC score | 0.076149 (rank : 71) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q92527, Q96QN1, Q9UDM3 | Gene names | ANKRD7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 7 (Testis-specific protein TSA806). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.074263 (rank : 72) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANFY1_MOUSE
|
||||||
| NC score | 0.074033 (rank : 73) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.073149 (rank : 74) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.072913 (rank : 75) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
ASZ1_HUMAN
|
||||||
| NC score | 0.072848 (rank : 76) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WWH4 | Gene names | ASZ1, GASZ | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat, SAM and basic leucine zipper domain-containing protein 1 (Germ cell-specific ankyrin, SAM and basic leucine zipper domain- containing protein). | |||||
|
ASB1_MOUSE
|
||||||
| NC score | 0.072806 (rank : 77) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9WV74 | Gene names | Asb1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 1 (ASB-1). | |||||
|
ANR29_HUMAN
|
||||||
| NC score | 0.072690 (rank : 78) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N6D5, Q6ZWE8, Q96LU9 | Gene names | ANKRD29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 29. | |||||
|
GABP2_MOUSE
|
||||||
| NC score | 0.072309 (rank : 79) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q00420, Q00421, Q8BS31, Q91YZ0, Q9QVV2 | Gene names | Gabpb2, Gabpb, Gabpb1 | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1). | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.072300 (rank : 80) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.072270 (rank : 81) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.071070 (rank : 82) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
POT14_HUMAN
|
||||||
| NC score | 0.069849 (rank : 83) | θ value | 1.06291 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
ANR15_HUMAN
|
||||||
| NC score | 0.069587 (rank : 84) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q14678, Q5W0W0, Q8IY65, Q8WX74 | Gene names | ANKRD15, KANK, KIAA0172 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 15 (Kidney ankyrin repeat- containing protein). | |||||
|
ANR28_MOUSE
|
||||||
| NC score | 0.069572 (rank : 85) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
GABP2_HUMAN
|
||||||
| NC score | 0.068961 (rank : 86) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
AN13A_HUMAN
|
||||||
| NC score | 0.068589 (rank : 87) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8IZ07, O60736 | Gene names | ANKRD13A, ANKRD13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13A (Protein KE03). | |||||
|
ANKR1_MOUSE
|
||||||
| NC score | 0.068282 (rank : 88) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANR28_HUMAN
|
||||||
| NC score | 0.068193 (rank : 89) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ASB15_HUMAN
|
||||||
| NC score | 0.067968 (rank : 90) | θ value | 0.125558 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8WXK1 | Gene names | ASB15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
MIB1_MOUSE
|
||||||
| NC score | 0.067151 (rank : 91) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
MIB1_HUMAN
|
||||||
| NC score | 0.067045 (rank : 92) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.066860 (rank : 93) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
ASB2_HUMAN
|
||||||
| NC score | 0.065944 (rank : 94) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96Q27, Q9NSU5, Q9Y567 | Gene names | ASB2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 2 (ASB-2). | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.065347 (rank : 95) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
BARD1_HUMAN
|
||||||
| NC score | 0.065328 (rank : 96) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.065062 (rank : 97) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.064793 (rank : 98) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
BCL3_HUMAN
|
||||||
| NC score | 0.064126 (rank : 99) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
ANR19_HUMAN
|
||||||
| NC score | 0.063050 (rank : 100) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H560 | Gene names | ANKRD19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 19. | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.062775 (rank : 101) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
ASB2_MOUSE
|
||||||
| NC score | 0.062339 (rank : 102) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8K0L0, Q9CTH4, Q9WV73 | Gene names | Asb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 2 (ASB-2). | |||||
|
ASB1_HUMAN
|
||||||
| NC score | 0.062307 (rank : 103) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y576, Q9ULS4 | Gene names | ASB1, KIAA1146 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 1 (ASB-1). | |||||
|
FANK1_HUMAN
|
||||||
| NC score | 0.062021 (rank : 104) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8TC84, Q6UXY9, Q6X7T6 | Gene names | FANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type 3 and ankyrin repeat domains protein 1. | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.061928 (rank : 105) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.061145 (rank : 106) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
PA2G6_MOUSE
|
||||||
| NC score | 0.061136 (rank : 107) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
AN36A_HUMAN
|
||||||
| NC score | 0.059981 (rank : 108) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6UX02, Q86X62 | Gene names | ANKRD36, ANKRD36A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 36A. | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.059688 (rank : 109) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
ASB15_MOUSE
|
||||||
| NC score | 0.059516 (rank : 110) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8VHS6 | Gene names | Asb15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.057915 (rank : 111) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CT012_MOUSE
|
||||||
| NC score | 0.056796 (rank : 112) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8C008, Q8C060 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C20orf12 homolog. | |||||
|
ASB14_MOUSE
|
||||||
| NC score | 0.056444 (rank : 113) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8VHS7 | Gene names | Asb14 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 14 (ASB-14). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.055443 (rank : 114) | θ value | 0.0330416 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.055061 (rank : 115) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
POT22_HUMAN
|
||||||
| NC score | 0.054919 (rank : 116) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6S545 | Gene names | ACTBL1, POTE22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 22. | |||||
|
ANFY1_HUMAN
|
||||||
| NC score | 0.054859 (rank : 117) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | 0.054462 (rank : 118) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
FANK1_MOUSE
|
||||||
| NC score | 0.054353 (rank : 119) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9DAM9, Q9CUA7 | Gene names | Fank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type 3 and ankyrin repeat domains 1 protein (Germ cell- specific gene 1 protein) (GSG1). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.054300 (rank : 120) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
ASB7_HUMAN
|
||||||
| NC score | 0.054285 (rank : 121) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H672, Q7Z4S3 | Gene names | ASB7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB7_MOUSE
|
||||||
| NC score | 0.054247 (rank : 122) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q91ZU0 | Gene names | Asb7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
PA2G6_HUMAN
|
||||||
| NC score | 0.054227 (rank : 123) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
ANR18_HUMAN
|
||||||
| NC score | 0.053983 (rank : 124) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
MIB2_HUMAN
|
||||||
| NC score | 0.053922 (rank : 125) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
NFKB2_MOUSE
|
||||||
| NC score | 0.053746 (rank : 126) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
PSD10_HUMAN
|
||||||
| NC score | 0.053686 (rank : 127) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
MIB2_MOUSE
|
||||||
| NC score | 0.053506 (rank : 128) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
ANKR1_HUMAN
|
||||||
| NC score | 0.053486 (rank : 129) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
IKBE_MOUSE
|
||||||
| NC score | 0.053429 (rank : 130) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
ANRA2_MOUSE
|
||||||
| NC score | 0.053271 (rank : 131) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ASB6_HUMAN
|
||||||
| NC score | 0.053176 (rank : 132) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NWX5 | Gene names | ASB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
ANR25_MOUSE
|
||||||
| NC score | 0.053147 (rank : 133) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
ANR32_MOUSE
|
||||||
| NC score | 0.053103 (rank : 134) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8R3P9, Q5XK30, Q8BHY5, Q921Y9 | Gene names | Ankrd32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 32. | |||||
|
ANR16_HUMAN
|
||||||
| NC score | 0.053067 (rank : 135) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6P6B7, Q9NT01 | Gene names | ANKRD16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 16. | |||||
|
ANRA2_HUMAN
|
||||||
| NC score | 0.052912 (rank : 136) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
IKBA_HUMAN
|
||||||
| NC score | 0.052761 (rank : 137) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P25963 | Gene names | NFKBIA, IKBA, MAD3, NFKBI | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor alpha (Major histocompatibility complex enhancer- binding protein MAD3) (I-kappa-B-alpha) (IkappaBalpha) (IKB-alpha). | |||||
|
IKBA_MOUSE
|
||||||
| NC score | 0.052643 (rank : 138) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Z1E3, Q80ZX5 | Gene names | Nfkbia, Ikba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B inhibitor alpha (I-kappa-B-alpha) (IkappaBalpha) (IKB- alpha). | |||||
|
ANKR2_HUMAN
|
||||||
| NC score | 0.052575 (rank : 139) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9GZV1, Q5T456, Q8WUD7 | Gene names | ANKRD2, ARPP | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (hArpp). | |||||
|
EF2K_MOUSE
|
||||||
| NC score | 0.052500 (rank : 140) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
ASB6_MOUSE
|
||||||
| NC score | 0.052272 (rank : 141) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q91ZU1, Q8VEC6 | Gene names | Asb6 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.051947 (rank : 142) | θ value | 0.0330416 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.051687 (rank : 143) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
ANKR2_MOUSE
|
||||||
| NC score | 0.051685 (rank : 144) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9WV06 | Gene names | Ankrd2, Arpp | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.051671 (rank : 145) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
IKBB_HUMAN
|
||||||
| NC score | 0.051348 (rank : 146) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
ANR49_MOUSE
|
||||||
| NC score | 0.051164 (rank : 147) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8VE42, Q9Z2S1 | Gene names | Ankrd49, Fgif, Gbif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor) (Fetal globin-increasing factor). | |||||
|
IKBB_MOUSE
|
||||||
| NC score | 0.051004 (rank : 148) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
ANR46_HUMAN
|
||||||
| NC score | 0.050958 (rank : 149) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q86W74, Q6P9B7 | Gene names | ANKRD46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
CLPB_HUMAN
|
||||||
| NC score | 0.050937 (rank : 150) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H078, Q8ND11, Q9H8Y0 | Gene names | CLPB, SKD3 | |||
|
Domain Architecture |

|
|||||
| Description | Caseinolytic peptidase B protein homolog (Suppressor of potassium transport defect 3). | |||||
|
ANR22_HUMAN
|
||||||
| NC score | 0.050689 (rank : 151) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
ASB8_HUMAN
|
||||||
| NC score | 0.050540 (rank : 152) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
EF2K_HUMAN
|
||||||
| NC score | 0.050527 (rank : 153) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
PSD10_MOUSE
|
||||||
| NC score | 0.050305 (rank : 154) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.049368 (rank : 155) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.046856 (rank : 156) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.045170 (rank : 157) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.043550 (rank : 158) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CT026_HUMAN
|
||||||
| NC score | 0.043068 (rank : 159) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NHU2, Q5TE18, Q8N5R9, Q96M59, Q9BQL2, Q9H127, Q9H128, Q9NQH4, Q9UFV8, Q9Y4V7 | Gene names | C20orf26 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf26. | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.042916 (rank : 160) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.041955 (rank : 161) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.041901 (rank : 162) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
RIPK4_HUMAN
|
||||||
| NC score | 0.041723 (rank : 163) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1094 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P57078, Q96KH0 | Gene names | RIPK4, ANKRD3, DIK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 4 (EC 2.7.11.1) (Ankyrin repeat domain protein 3) (PKC-delta-interacting protein kinase). | |||||
|
ANKK1_HUMAN
|
||||||
| NC score | 0.041254 (rank : 164) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1075 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8NFD2 | Gene names | ANKK1, PKK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1) (Protein kinase PKK2) (X-kinase) (SgK288). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.038705 (rank : 165) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.036212 (rank : 166) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
ANKK1_MOUSE
|
||||||
| NC score | 0.036062 (rank : 167) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8BZ25 | Gene names | Ankk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.034900 (rank : 168) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
MPP10_MOUSE
|
||||||
| NC score | 0.031948 (rank : 169) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.027609 (rank : 170) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
TR143_MOUSE
|
||||||
| NC score | 0.026103 (rank : 171) | θ value | 0.813845 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQB9, Q2NL43 | Gene names | Tas2r143, T2r36, Tas2r43 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 143 (T2R143) (Taste receptor type 2 member 43) (T2R43) (mT2R36). | |||||
|
ANKF1_HUMAN
|
||||||
| NC score | 0.019733 (rank : 172) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N957 | Gene names | ANKFN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and fibronectin type-III domain-containing protein 1. | |||||
|
CTNS_MOUSE
|
||||||
| NC score | 0.019310 (rank : 173) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57757 | Gene names | Ctns | |||
|
Domain Architecture |

|
|||||
| Description | Cystinosin. | |||||
|
CDC37_MOUSE
|
||||||
| NC score | 0.018347 (rank : 174) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.017735 (rank : 175) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
VPS18_MOUSE
|
||||||
| NC score | 0.017024 (rank : 176) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R307, Q8BGV6, Q8BZX6, Q8C126 | Gene names | Vps18 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18. | |||||
|
CDC37_HUMAN
|
||||||
| NC score | 0.016075 (rank : 177) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
ASB17_MOUSE
|
||||||
| NC score | 0.012271 (rank : 178) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHP9, Q9CUF9 | Gene names | Asb17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 17 (ASB-17). | |||||
|
DOPO_HUMAN
|
||||||
| NC score | 0.011835 (rank : 179) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09172, Q5T381, Q96AG2 | Gene names | DBH | |||
|
Domain Architecture |

|
|||||
| Description | Dopamine beta-hydroxylase precursor (EC 1.14.17.1) (Dopamine beta- monooxygenase). | |||||
|
RBBP8_HUMAN
|
||||||
| NC score | 0.011347 (rank : 180) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
PCDGC_HUMAN
|
||||||
| NC score | 0.009566 (rank : 181) | θ value | 0.163984 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60330, O15100, Q6UW70, Q9Y5D7 | Gene names | PCDHGA12, CDH21, FIB3, KIAA0588 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A12 precursor (PCDH-gamma-A12) (Cadherin-21) (Fibroblast cadherin 3). | |||||
|
PCDGB_HUMAN
|
||||||
| NC score | 0.008936 (rank : 182) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5H2, Q9Y5D8, Q9Y5D9 | Gene names | PCDHGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A11 precursor (PCDH-gamma-A11). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.008395 (rank : 183) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
PCDG7_HUMAN
|
||||||
| NC score | 0.007995 (rank : 184) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5G6, Q9Y5D0 | Gene names | PCDHGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A7 precursor (PCDH-gamma-A7). | |||||
|
NET4_HUMAN
|
||||||
| NC score | 0.007054 (rank : 185) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HB63, Q658K9, Q7L3F1, Q7L9D6, Q7Z5B6, Q9BZP1, Q9NT44, Q9P133 | Gene names | NTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin) (Hepar-derived netrin-like protein). | |||||
|
K0831_MOUSE
|
||||||
| NC score | 0.006916 (rank : 186) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
LZTS2_MOUSE
|
||||||
| NC score | 0.006802 (rank : 187) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.005238 (rank : 188) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.004000 (rank : 189) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.003992 (rank : 190) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.003836 (rank : 191) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.003713 (rank : 192) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.003557 (rank : 193) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.002161 (rank : 194) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.001012 (rank : 195) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SLK_HUMAN
|
||||||
| NC score | -0.003605 (rank : 196) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||