Please be patient as the page loads
|
TTP_MOUSE
|
||||||
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TTP_MOUSE
|
||||||
| θ value | 1.75107e-120 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
TTP_HUMAN
|
||||||
| θ value | 3.9282e-96 (rank : 2) | NC score | 0.974756 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |
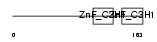
|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
TISD_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 3) | NC score | 0.857965 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
TISB_MOUSE
|
||||||
| θ value | 2.05525e-28 (rank : 4) | NC score | 0.856671 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |
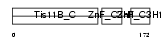
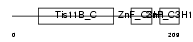
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
TISB_HUMAN
|
||||||
| θ value | 2.68423e-28 (rank : 5) | NC score | 0.855166 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |
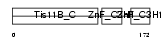
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISD_MOUSE
|
||||||
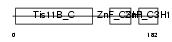
| θ value | 7.80994e-28 (rank : 6) | NC score | 0.866056 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
ZC3H5_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 7) | NC score | 0.323892 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ZC3H5_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 8) | NC score | 0.313333 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9C0B0 | Gene names | ZC3H5, KIAA1753, ZC3HDC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 9) | NC score | 0.300727 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 10) | NC score | 0.300833 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 11) | NC score | 0.305317 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MNAB_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 12) | NC score | 0.240986 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
MNAB_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 13) | NC score | 0.243977 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.299087 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 15) | NC score | 0.297430 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
DUS3L_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 16) | NC score | 0.249127 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
RC3H1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 17) | NC score | 0.244585 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 18) | NC score | 0.240474 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 19) | NC score | 0.272312 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 20) | NC score | 0.337151 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 21) | NC score | 0.034542 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 22) | NC score | 0.277060 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.185142 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
PAR12_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.103159 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.113318 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.266786 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.292106 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
DUS3L_MOUSE
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.192633 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91XI1, Q3TZQ1, Q7TT12 | Gene names | Dus3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
PAR12_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.104693 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.056802 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.045281 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.333086 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
MBN3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.105679 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
MBNL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.090484 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.249751 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.255429 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
CP007_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.045424 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.061680 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.186522 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.065134 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
SIA1A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.067182 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61092, Q06984 | Gene names | Siah1a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase SIAH1A (EC 6.3.2.-) (Seven in absentia homolog 1a) (Siah1a) (Siah-1a) (mSiah-1a). | |||||
|
SIAH1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.067142 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IUQ4, O43269, Q92880 | Gene names | SIAH1, HUMSIAH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase SIAH1 (EC 6.3.2.-) (Seven in absentia homolog 1) (Siah-1) (Siah-1a). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.025668 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
FA8_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.021404 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.220699 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.007420 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.288998 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.043704 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
R113A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.139846 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
R113B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.141712 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.051083 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
HXB13_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.011499 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70321, Q80Y12 | Gene names | Hoxb13 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B13. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.016341 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TESK2_MOUSE
|
||||||
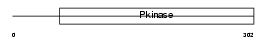
| θ value | 2.36792 (rank : 54) | NC score | 0.000126 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |
|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
CT174_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.040274 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
DCHS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.028112 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23738 | Gene names | Hdc | |||
|
Domain Architecture |

|
|||||
| Description | Histidine decarboxylase (EC 4.1.1.22) (HDC). | |||||
|
HXA11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.011389 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31270 | Gene names | HOXA11, HOX1I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1I). | |||||
|
HXA11_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.011383 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31311, Q3V026 | Gene names | Hoxa11, Hox-1.9, Hoxa-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1.9). | |||||
|
PCDA2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.003261 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5H9, O75287, Q9BTV3 | Gene names | PCDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 2 precursor (PCDH-alpha2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.032700 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
GAB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.016967 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
HXB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.006650 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14653 | Gene names | HOXB1, HOX2I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B1 (Hox-2I). | |||||
|
LYST_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.010155 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.025769 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.013327 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
IL3RB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.017255 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.007556 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.009563 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PRR3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.054923 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q811B5, Q91YI5 | Gene names | Prr3, Cat56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 3 (MHC class I region proline-rich protein CAT56). | |||||
|
ATS14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.003981 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
B4GT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.008032 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.013817 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
LECT1_MOUSE
|
||||||
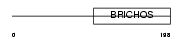
| θ value | 8.99809 (rank : 73) | NC score | 0.011878 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1F6, Q9CXU5 | Gene names | Lect1, Chmi | |||
|
Domain Architecture |
|
|||||
| Description | Chondromodulin-1 precursor (Chondromodulin-I) (ChM-I) (Leukocyte cell- derived chemotaxin 1) [Contains: Chondrosurfactant protein (CH-SP)]. | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.023306 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SH2D3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.008893 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
MBN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.068809 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
MBNL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.057331 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NR56, O43311, O43797 | Gene names | MBNL1, EXP, KIAA0428, MBNL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.75107e-120 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
TTP_HUMAN
|
||||||
| NC score | 0.974756 (rank : 2) | θ value | 3.9282e-96 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
TISD_MOUSE
|
||||||
| NC score | 0.866056 (rank : 3) | θ value | 7.80994e-28 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
TISD_HUMAN
|
||||||
| NC score | 0.857965 (rank : 4) | θ value | 5.40856e-29 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
TISB_MOUSE
|
||||||
| NC score | 0.856671 (rank : 5) | θ value | 2.05525e-28 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
TISB_HUMAN
|
||||||
| NC score | 0.855166 (rank : 6) | θ value | 2.68423e-28 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.337151 (rank : 7) | θ value | 0.00665767 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.333086 (rank : 8) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
ZC3H5_MOUSE
|
||||||
| NC score | 0.323892 (rank : 9) | θ value | 7.1131e-05 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ZC3H5_HUMAN
|
||||||
| NC score | 0.313333 (rank : 10) | θ value | 0.000461057 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9C0B0 | Gene names | ZC3H5, KIAA1753, ZC3HDC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.305317 (rank : 11) | θ value | 0.00134147 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.300833 (rank : 12) | θ value | 0.00102713 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.300727 (rank : 13) | θ value | 0.000786445 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.299087 (rank : 14) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.297430 (rank : 15) | θ value | 0.00509761 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.292106 (rank : 16) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.288998 (rank : 17) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.277060 (rank : 18) | θ value | 0.0148317 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.272312 (rank : 19) | θ value | 0.00665767 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.266786 (rank : 20) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.255429 (rank : 21) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.249751 (rank : 22) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
DUS3L_HUMAN
|
||||||
| NC score | 0.249127 (rank : 23) | θ value | 0.00509761 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
RC3H1_MOUSE
|
||||||
| NC score | 0.244585 (rank : 24) | θ value | 0.00509761 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
MNAB_MOUSE
|
||||||
| NC score | 0.243977 (rank : 25) | θ value | 0.00390308 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
MNAB_HUMAN
|
||||||
| NC score | 0.240986 (rank : 26) | θ value | 0.00390308 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.240474 (rank : 27) | θ value | 0.00665767 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.220699 (rank : 28) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
DUS3L_MOUSE
|
||||||
| NC score | 0.192633 (rank : 29) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91XI1, Q3TZQ1, Q7TT12 | Gene names | Dus3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.186522 (rank : 30) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.185142 (rank : 31) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
R113B_HUMAN
|
||||||
| NC score | 0.141712 (rank : 32) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
R113A_HUMAN
|
||||||
| NC score | 0.139846 (rank : 33) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.113318 (rank : 34) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
MBN3_HUMAN
|
||||||
| NC score | 0.105679 (rank : 35) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
PAR12_HUMAN
|
||||||
| NC score | 0.104693 (rank : 36) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PAR12_MOUSE
|
||||||
| NC score | 0.103159 (rank : 37) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
MBNL_MOUSE
|
||||||
| NC score | 0.090484 (rank : 38) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MBN3_MOUSE
|
||||||
| NC score | 0.068809 (rank : 39) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
SIA1A_MOUSE
|
||||||
| NC score | 0.067182 (rank : 40) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61092, Q06984 | Gene names | Siah1a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase SIAH1A (EC 6.3.2.-) (Seven in absentia homolog 1a) (Siah1a) (Siah-1a) (mSiah-1a). | |||||
|
SIAH1_HUMAN
|
||||||
| NC score | 0.067142 (rank : 41) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IUQ4, O43269, Q92880 | Gene names | SIAH1, HUMSIAH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase SIAH1 (EC 6.3.2.-) (Seven in absentia homolog 1) (Siah-1) (Siah-1a). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.065134 (rank : 42) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.061680 (rank : 43) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
MBNL_HUMAN
|
||||||
| NC score | 0.057331 (rank : 44) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NR56, O43311, O43797 | Gene names | MBNL1, EXP, KIAA0428, MBNL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.056802 (rank : 45) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
PRR3_MOUSE
|
||||||
| NC score | 0.054923 (rank : 46) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q811B5, Q91YI5 | Gene names | Prr3, Cat56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 3 (MHC class I region proline-rich protein CAT56). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.051083 (rank : 47) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
CP007_MOUSE
|
||||||
| NC score | 0.045424 (rank : 48) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.045281 (rank : 49) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.043704 (rank : 50) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.040274 (rank : 51) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.034542 (rank : 52) | θ value | 0.0148317 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.032700 (rank : 53) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DCHS_MOUSE
|
||||||
| NC score | 0.028112 (rank : 54) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23738 | Gene names | Hdc | |||
|
Domain Architecture |

|
|||||
| Description | Histidine decarboxylase (EC 4.1.1.22) (HDC). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.025769 (rank : 55) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.025668 (rank : 56) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.023306 (rank : 57) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.021404 (rank : 58) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
IL3RB_HUMAN
|
||||||
| NC score | 0.017255 (rank : 59) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.016967 (rank : 60) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.016341 (rank : 61) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.013817 (rank : 62) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.013327 (rank : 63) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
LECT1_MOUSE
|
||||||
| NC score | 0.011878 (rank : 64) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1F6, Q9CXU5 | Gene names | Lect1, Chmi | |||
|
Domain Architecture |

|
|||||
| Description | Chondromodulin-1 precursor (Chondromodulin-I) (ChM-I) (Leukocyte cell- derived chemotaxin 1) [Contains: Chondrosurfactant protein (CH-SP)]. | |||||
|
HXB13_MOUSE
|
||||||
| NC score | 0.011499 (rank : 65) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70321, Q80Y12 | Gene names | Hoxb13 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B13. | |||||
|
HXA11_HUMAN
|
||||||
| NC score | 0.011389 (rank : 66) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31270 | Gene names | HOXA11, HOX1I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1I). | |||||
|
HXA11_MOUSE
|
||||||
| NC score | 0.011383 (rank : 67) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31311, Q3V026 | Gene names | Hoxa11, Hox-1.9, Hoxa-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1.9). | |||||
|
LYST_MOUSE
|
||||||
| NC score | 0.010155 (rank : 68) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.009563 (rank : 69) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
SH2D3_HUMAN
|
||||||
| NC score | 0.008893 (rank : 70) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.008032 (rank : 71) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.007556 (rank : 72) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.007420 (rank : 73) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
HXB1_HUMAN
|
||||||
| NC score | 0.006650 (rank : 74) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14653 | Gene names | HOXB1, HOX2I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B1 (Hox-2I). | |||||
|
ATS14_HUMAN
|
||||||
| NC score | 0.003981 (rank : 75) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
PCDA2_HUMAN
|
||||||
| NC score | 0.003261 (rank : 76) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5H9, O75287, Q9BTV3 | Gene names | PCDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 2 precursor (PCDH-alpha2). | |||||
|
TESK2_MOUSE
|
||||||
| NC score | 0.000126 (rank : 77) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||