Please be patient as the page loads
|
B4GT1_HUMAN
|
||||||
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
B4GT1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 168 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982690 (rank : 2) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_HUMAN
|
||||||
| θ value | 1.13764e-111 (rank : 3) | NC score | 0.973874 (rank : 4) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| θ value | 4.04623e-109 (rank : 4) | NC score | 0.974704 (rank : 3) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_HUMAN
|
||||||
| θ value | 4.20666e-90 (rank : 5) | NC score | 0.965199 (rank : 6) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_MOUSE
|
||||||
| θ value | 9.37149e-90 (rank : 6) | NC score | 0.966982 (rank : 5) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_MOUSE
|
||||||
| θ value | 2.64714e-76 (rank : 7) | NC score | 0.963278 (rank : 7) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_HUMAN
|
||||||
| θ value | 2.92676e-75 (rank : 8) | NC score | 0.961573 (rank : 8) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT5_MOUSE
|
||||||
| θ value | 8.83269e-64 (rank : 9) | NC score | 0.936458 (rank : 9) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_HUMAN
|
||||||
| θ value | 5.7252e-63 (rank : 10) | NC score | 0.936192 (rank : 10) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT6_HUMAN
|
||||||
| θ value | 5.0155e-59 (rank : 11) | NC score | 0.933474 (rank : 11) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| θ value | 4.24587e-58 (rank : 12) | NC score | 0.932496 (rank : 12) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT7_MOUSE
|
||||||
| θ value | 7.54701e-31 (rank : 13) | NC score | 0.875430 (rank : 13) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT7_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 14) | NC score | 0.868129 (rank : 14) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
CHSS1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.157950 (rank : 15) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.151515 (rank : 16) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.130862 (rank : 17) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.014453 (rank : 73) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
GBF1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 19) | NC score | 0.026154 (rank : 39) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
TM55B_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.033984 (rank : 29) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86T03, Q86U09, Q8WUC0, Q9BU67, Q9NSU8 | Gene names | TMEM55B, C14orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.041542 (rank : 27) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.022873 (rank : 47) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.023573 (rank : 46) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.009280 (rank : 117) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
RANB9_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.031973 (rank : 31) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.018206 (rank : 63) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
BRD3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.021273 (rank : 49) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
DAZP2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.053596 (rank : 25) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.053781 (rank : 24) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
GP178_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.041803 (rank : 26) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2C4, Q5VTU1 | Gene names | GPR178, KIAA1423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR178. | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.004547 (rank : 148) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
YTHD1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.021064 (rank : 50) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59326, Q3T9E2 | Gene names | Ythdf1 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1 homolog) (DACA-1 homolog). | |||||
|
BSPRY_MOUSE
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.014399 (rank : 74) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80YW5, Q3TU74, Q8BZF0, Q99KV7, Q9ER70 | Gene names | Bspry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B box and SPRY domain-containing protein. | |||||
|
MNT_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.032559 (rank : 30) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
SPAG8_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.041532 (rank : 28) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99932, Q12937, Q5TCV8, Q8WWB4 | Gene names | SPAG8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 8 (Sperm membrane protein 1) (SMP-1) (HSD-1) (Sperm membrane protein BS-84). | |||||
|
VAX1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.012349 (rank : 89) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q2NKI2, O88880 | Gene names | Vax1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
WASL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.020385 (rank : 54) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CGAT2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.084223 (rank : 21) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.004682 (rank : 147) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.016533 (rank : 70) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.019046 (rank : 59) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
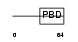
BORG5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.024730 (rank : 42) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |
|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.005824 (rank : 140) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
MTF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.000349 (rank : 163) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.014241 (rank : 76) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PYGO2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.029565 (rank : 33) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
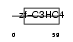
RING1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.019780 (rank : 55) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |
|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.024149 (rank : 44) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
VAX1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.011584 (rank : 96) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SQQ9, Q6ZSX0 | Gene names | VAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.019076 (rank : 58) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.009793 (rank : 112) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
GMIP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.011598 (rank : 95) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.020739 (rank : 53) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HES6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.029112 (rank : 34) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.010857 (rank : 102) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
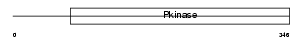
STK39_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.000175 (rank : 164) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |
|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
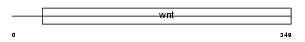
WNT3A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.012448 (rank : 86) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27467 | Gene names | Wnt3a, Wnt-3a | |||
|
Domain Architecture |
|
|||||
| Description | Protein Wnt-3a precursor. | |||||
|
ECM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.025844 (rank : 40) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.020991 (rank : 51) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HES6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.027756 (rank : 37) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
HNF3A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.012125 (rank : 92) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55317, Q9H2A0 | Gene names | FOXA1, HNF3A, TCF3A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
HNF3A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.012428 (rank : 87) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35582, Q61108 | Gene names | Foxa1, Hnf3a, Tcf-3a, Tcf3a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
RUSC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.023840 (rank : 45) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.028614 (rank : 35) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.020936 (rank : 52) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.003505 (rank : 154) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ACM4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.000476 (rank : 162) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32211, Q64056 | Gene names | Chrm4, Chrm-4 | |||
|
Domain Architecture |
|
|||||
| Description | Muscarinic acetylcholine receptor M4 (Mm4 mAChR). | |||||
|
CGAT1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.089009 (rank : 18) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CN004_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.025107 (rank : 41) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
DIAP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.012472 (rank : 85) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.008601 (rank : 123) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.014108 (rank : 78) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RING1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.018078 (rank : 65) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
WNT3A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.011419 (rank : 98) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56704, Q969P2 | Gene names | WNT3A | |||
|
Domain Architecture |
|
|||||
| Description | Protein Wnt-3a precursor. | |||||
|
CGAT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.086137 (rank : 20) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.013131 (rank : 81) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
DOK7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.017277 (rank : 67) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q18PE1, Q6P6A6, Q86XG5, Q8N2J3, Q8NBC1 | Gene names | DOK7, C4orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.019541 (rank : 56) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
JIP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.011136 (rank : 100) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.018461 (rank : 60) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.026555 (rank : 38) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
OTX1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.006128 (rank : 137) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P32242, Q53TG6 | Gene names | OTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
PDLI5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.008750 (rank : 122) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.003728 (rank : 153) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZN219_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.002202 (rank : 160) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.004267 (rank : 150) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CGAT2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.086685 (rank : 19) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CT055_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.018345 (rank : 62) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQ89 | Gene names | C20orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf55. | |||||
|
EMID2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.009704 (rank : 113) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96A83 | Gene names | EMID2, COL26A1, EMU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.008865 (rank : 121) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
HNF3G_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.009199 (rank : 118) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55318, Q9UMW9 | Gene names | FOXA3, HNF3G, TCF3G | |||
|
Domain Architecture |
|
|||||
| Description | Hepatocyte nuclear factor 3-gamma (HNF-3G) (Forkhead box protein A3) (Fork head-related protein FKH H3). | |||||
|
JAD1C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.010434 (rank : 109) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
M3K12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | -0.000563 (rank : 165) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.014695 (rank : 72) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.012837 (rank : 84) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.027969 (rank : 36) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.012315 (rank : 90) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.006494 (rank : 134) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.010806 (rank : 103) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CABL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.011133 (rank : 101) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESJ1, Q9EPR8 | Gene names | Cables1, Cables | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.005944 (rank : 139) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.003829 (rank : 151) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
CN004_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.022821 (rank : 48) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
CXA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.004307 (rank : 149) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6H8, Q9H537 | Gene names | GJA3 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-3 protein (Connexin-46) (Cx46). | |||||
|
EFCB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.010708 (rank : 104) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VUJ9, Q59G23, Q9BS36 | Gene names | EFCAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
GGN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.014191 (rank : 77) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.019290 (rank : 57) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
K1196_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.006716 (rank : 132) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.010700 (rank : 105) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NKX61_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.005297 (rank : 145) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.012191 (rank : 91) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.011893 (rank : 94) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
ZSWM5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.014350 (rank : 75) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P217, Q5SXQ9 | Gene names | ZSWIM5, KIAA1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
ANR47_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.005685 (rank : 142) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NY19, Q6NZI1, Q6ZQR3, Q8IUV2 | Gene names | ANKRD47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.006813 (rank : 131) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BTBD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.003795 (rank : 152) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BX70, O60418, O75248, Q7Z5W0, Q96SX8, Q9NPS1, Q9NX81 | Gene names | BTBD2 | |||
|
Domain Architecture |
|
|||||
| Description | BTB/POZ domain-containing protein 2. | |||||
|
CO1A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.006635 (rank : 133) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.008942 (rank : 120) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.010358 (rank : 110) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
HNRL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.010630 (rank : 106) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
M3K11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | -0.000997 (rank : 168) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||
|
MBRL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.012375 (rank : 88) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CIV2, Q68EE1, Q6KAN7, Q7TS84, Q8CIV1, Q99K29 | Gene names | ORF61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membralin. | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.011564 (rank : 97) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PLS3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.009834 (rank : 111) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PRR13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.018091 (rank : 64) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.005541 (rank : 144) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SP5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.000539 (rank : 161) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.016738 (rank : 69) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.018008 (rank : 66) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
ZSWM5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.013687 (rank : 79) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TC6 | Gene names | Zswim5, Kiaa1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
CAMKV_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | -0.000751 (rank : 166) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3UHL1, Q8VD20 | Gene names | Camkv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CCD1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.009443 (rank : 115) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.012849 (rank : 83) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.009349 (rank : 116) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
KCAB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.006088 (rank : 138) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43448 | Gene names | KCNAB3, KCNA3B | |||
|
Domain Architecture |
|
|||||
| Description | Voltage-gated potassium channel subunit beta-3 (K(+) channel subunit beta-3) (Kv-beta-3). | |||||
|
PXK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.007867 (rank : 129) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
RANB9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.024502 (rank : 43) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P69566 | Gene names | Ranbp9, Ranbpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (Ran-binding protein M) (RanBPM) (B cell antigen receptor Ig beta-associated protein 1) (IBAP-1). | |||||
|
SCMH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.005824 (rank : 141) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96GD3, Q5VT76, Q6IAJ4, Q8WU48, Q9UKM5, Q9UKM6 | Gene names | SCMH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.013039 (rank : 82) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SSBP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.010474 (rank : 108) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P81877, Q8N2Q2, Q9BWW6, Q9Y4T7 | Gene names | SSBP2, SSDP2 | |||
|
Domain Architecture |
|
|||||
| Description | Single-stranded DNA-binding protein 2 (Sequence-specific single- stranded-DNA-binding protein 2). | |||||
|
SSBP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.010493 (rank : 107) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CYZ8, Q99LX9 | Gene names | Ssbp2, Ssdp2 | |||
|
Domain Architecture |
|
|||||
| Description | Single-stranded DNA-binding protein 2 (Sequence-specific single- stranded-DNA-binding protein 2). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.007402 (rank : 130) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.008119 (rank : 124) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
ACM4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | -0.000939 (rank : 167) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08173 | Gene names | CHRM4 | |||
|
Domain Architecture |
|
|||||
| Description | Muscarinic acetylcholine receptor M4. | |||||
|
CBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.017231 (rank : 68) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.005606 (rank : 143) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.030462 (rank : 32) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.009697 (rank : 114) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.002862 (rank : 157) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
FBLI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.006133 (rank : 136) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WUP2, Q5VVE0, Q5VVE1, Q8IX23, Q96T00, Q9UFD6 | Gene names | FBLIM1, FBLP1 | |||
|
Domain Architecture |
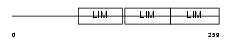
|
|||||
| Description | Filamin-binding LIM protein 1 (Filamin-binding LIM protein 1) (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin). | |||||
|
FOXF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.006209 (rank : 135) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12947, Q9UQ85 | Gene names | FOXF2, FKHL6, FREAC2 | |||
|
Domain Architecture |
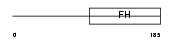
|
|||||
| Description | Forkhead box protein F2 (Forkhead-related protein FKHL6) (Forkhead- related transcription factor 2) (FREAC-2) (Forkhead-related activator 2). | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.018452 (rank : 61) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HXA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.003341 (rank : 156) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
JIP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.009005 (rank : 119) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | -0.001818 (rank : 170) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.008103 (rank : 125) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NEUR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.015563 (rank : 71) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ49, Q9NQE1 | Gene names | NEU3 | |||
|
Domain Architecture |
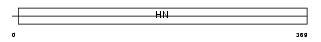
|
|||||
| Description | Sialidase-3 (EC 3.2.1.18) (Membrane sialidase) (Ganglioside sialidase) (N-acetyl-alpha-neuraminidase 3). | |||||
|
OST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.002769 (rank : 158) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y661, Q5QI42, Q8NDC2 | Gene names | HS3ST4, 3OST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 4 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4) (Heparan sulfate 3-O-sulfotransferase 4) (h3-OST-4). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.012107 (rank : 93) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.002501 (rank : 159) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.004926 (rank : 146) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.013664 (rank : 80) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.011195 (rank : 99) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
TISD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.007913 (rank : 128) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |
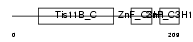
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
TTP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.008032 (rank : 126) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
WASF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.008018 (rank : 127) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.003426 (rank : 155) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | -0.000999 (rank : 169) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.070994 (rank : 22) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.062922 (rank : 23) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 168 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_MOUSE
|
||||||
| NC score | 0.982690 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| NC score | 0.974704 (rank : 3) | θ value | 4.04623e-109 (rank : 4) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_HUMAN
|
||||||
| NC score | 0.973874 (rank : 4) | θ value | 1.13764e-111 (rank : 3) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_MOUSE
|
||||||
| NC score | 0.966982 (rank : 5) | θ value | 9.37149e-90 (rank : 6) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_HUMAN
|
||||||
| NC score | 0.965199 (rank : 6) | θ value | 4.20666e-90 (rank : 5) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_MOUSE
|
||||||
| NC score | 0.963278 (rank : 7) | θ value | 2.64714e-76 (rank : 7) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_HUMAN
|
||||||
| NC score | 0.961573 (rank : 8) | θ value | 2.92676e-75 (rank : 8) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT5_MOUSE
|
||||||
| NC score | 0.936458 (rank : 9) | θ value | 8.83269e-64 (rank : 9) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_HUMAN
|
||||||
| NC score | 0.936192 (rank : 10) | θ value | 5.7252e-63 (rank : 10) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT6_HUMAN
|
||||||
| NC score | 0.933474 (rank : 11) | θ value | 5.0155e-59 (rank : 11) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| NC score | 0.932496 (rank : 12) | θ value | 4.24587e-58 (rank : 12) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT7_MOUSE
|
||||||
| NC score | 0.875430 (rank : 13) | θ value | 7.54701e-31 (rank : 13) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT7_HUMAN
|
||||||
| NC score | 0.868129 (rank : 14) | θ value | 1.08979e-29 (rank : 14) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
CHSS1_HUMAN
|
||||||
| NC score | 0.157950 (rank : 15) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.151515 (rank : 16) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.130862 (rank : 17) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CGAT1_MOUSE
|
||||||
| NC score | 0.089009 (rank : 18) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_MOUSE
|
||||||
| NC score | 0.086685 (rank : 19) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT1_HUMAN
|
||||||
| NC score | 0.086137 (rank : 20) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| NC score | 0.084223 (rank : 21) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.070994 (rank : 22) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.062922 (rank : 23) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
DAZP2_MOUSE
|
||||||
| NC score | 0.053781 (rank : 24) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
DAZP2_HUMAN
|
||||||
| NC score | 0.053596 (rank : 25) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
GP178_HUMAN
|
||||||
| NC score | 0.041803 (rank : 26) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2C4, Q5VTU1 | Gene names | GPR178, KIAA1423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR178. | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.041542 (rank : 27) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
SPAG8_HUMAN
|
||||||
| NC score | 0.041532 (rank : 28) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99932, Q12937, Q5TCV8, Q8WWB4 | Gene names | SPAG8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 8 (Sperm membrane protein 1) (SMP-1) (HSD-1) (Sperm membrane protein BS-84). | |||||
|
TM55B_HUMAN
|
||||||
| NC score | 0.033984 (rank : 29) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86T03, Q86U09, Q8WUC0, Q9BU67, Q9NSU8 | Gene names | TMEM55B, C14orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.032559 (rank : 30) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
RANB9_HUMAN
|
||||||
| NC score | 0.031973 (rank : 31) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.030462 (rank : 32) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
PYGO2_HUMAN
|
||||||
| NC score | 0.029565 (rank : 33) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
HES6_MOUSE
|
||||||
| NC score | 0.029112 (rank : 34) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.028614 (rank : 35) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.027969 (rank : 36) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
HES6_HUMAN
|
||||||
| NC score | 0.027756 (rank : 37) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.026555 (rank : 38) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
GBF1_HUMAN
|
||||||
| NC score | 0.026154 (rank : 39) | θ value | 0.0736092 (rank : 19) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
ECM1_HUMAN
|
||||||
| NC score | 0.025844 (rank : 40) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.025107 (rank : 41) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
BORG5_HUMAN
|
||||||
| NC score | 0.024730 (rank : 42) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
RANB9_MOUSE
|
||||||
| NC score | 0.024502 (rank : 43) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P69566 | Gene names | Ranbp9, Ranbpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (Ran-binding protein M) (RanBPM) (B cell antigen receptor Ig beta-associated protein 1) (IBAP-1). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.024149 (rank : 44) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
RUSC2_HUMAN
|
||||||
| NC score | 0.023840 (rank : 45) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.023573 (rank : 46) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.022873 (rank : 47) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
CN004_HUMAN
|
||||||
| NC score | 0.022821 (rank : 48) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
BRD3_HUMAN
|
||||||
| NC score | 0.021273 (rank : 49) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
YTHD1_MOUSE
|
||||||
| NC score | 0.021064 (rank : 50) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59326, Q3T9E2 | Gene names | Ythdf1 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1 homolog) (DACA-1 homolog). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.020991 (rank : 51) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.020936 (rank : 52) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.020739 (rank : 53) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.020385 (rank : 54) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
RING1_HUMAN
|
||||||
| NC score | 0.019780 (rank : 55) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.019541 (rank : 56) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.019290 (rank : 57) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.019076 (rank : 58) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.019046 (rank : 59) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.018461 (rank : 60) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.018452 (rank : 61) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
CT055_HUMAN
|
||||||
| NC score | 0.018345 (rank : 62) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQ89 | Gene names | C20orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf55. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.018206 (rank : 63) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.018091 (rank : 64) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
RING1_MOUSE
|
||||||
| NC score | 0.018078 (rank : 65) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.018008 (rank : 66) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
DOK7_HUMAN
|
||||||
| NC score | 0.017277 (rank : 67) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q18PE1, Q6P6A6, Q86XG5, Q8N2J3, Q8NBC1 | Gene names | DOK7, C4orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.017231 (rank : 68) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.016738 (rank : 69) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.016533 (rank : 70) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NEUR3_HUMAN
|
||||||
| NC score | 0.015563 (rank : 71) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ49, Q9NQE1 | Gene names | NEU3 | |||
|
Domain Architecture |

|
|||||
| Description | Sialidase-3 (EC 3.2.1.18) (Membrane sialidase) (Ganglioside sialidase) (N-acetyl-alpha-neuraminidase 3). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.014695 (rank : 72) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.014453 (rank : 73) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
BSPRY_MOUSE
|
||||||
| NC score | 0.014399 (rank : 74) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80YW5, Q3TU74, Q8BZF0, Q99KV7, Q9ER70 | Gene names | Bspry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B box and SPRY domain-containing protein. | |||||
|
ZSWM5_HUMAN
|
||||||
| NC score | 0.014350 (rank : 75) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P217, Q5SXQ9 | Gene names | ZSWIM5, KIAA1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.014241 (rank : 76) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.014191 (rank : 77) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.014108 (rank : 78) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ZSWM5_MOUSE
|
||||||
| NC score | 0.013687 (rank : 79) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TC6 | Gene names | Zswim5, Kiaa1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.013664 (rank : 80) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.013131 (rank : 81) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.013039 (rank : 82) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.012849 (rank : 83) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.012837 (rank : 84) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
DIAP2_MOUSE
|
||||||
| NC score | 0.012472 (rank : 85) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
WNT3A_MOUSE
|
||||||
| NC score | 0.012448 (rank : 86) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27467 | Gene names | Wnt3a, Wnt-3a | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-3a precursor. | |||||
|
HNF3A_MOUSE
|
||||||
| NC score | 0.012428 (rank : 87) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35582, Q61108 | Gene names | Foxa1, Hnf3a, Tcf-3a, Tcf3a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
MBRL_MOUSE
|
||||||
| NC score | 0.012375 (rank : 88) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CIV2, Q68EE1, Q6KAN7, Q7TS84, Q8CIV1, Q99K29 | Gene names | ORF61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membralin. | |||||
|
VAX1_MOUSE
|
||||||
| NC score | 0.012349 (rank : 89) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q2NKI2, O88880 | Gene names | Vax1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.012315 (rank : 90) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.012191 (rank : 91) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
HNF3A_HUMAN
|
||||||
| NC score | 0.012125 (rank : 92) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55317, Q9H2A0 | Gene names | FOXA1, HNF3A, TCF3A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.012107 (rank : 93) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.011893 (rank : 94) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.011598 (rank : 95) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
VAX1_HUMAN
|
||||||
| NC score | 0.011584 (rank : 96) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SQQ9, Q6ZSX0 | Gene names | VAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.011564 (rank : 97) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
WNT3A_HUMAN
|
||||||
| NC score | 0.011419 (rank : 98) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56704, Q969P2 | Gene names | WNT3A | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-3a precursor. | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.011195 (rank : 99) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
JIP4_HUMAN
|
||||||
| NC score | 0.011136 (rank : 100) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
CABL1_MOUSE
|
||||||
| NC score | 0.011133 (rank : 101) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESJ1, Q9EPR8 | Gene names | Cables1, Cables | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.010857 (rank : 102) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.010806 (rank : 103) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
EFCB2_HUMAN
|
||||||
| NC score | 0.010708 (rank : 104) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VUJ9, Q59G23, Q9BS36 | Gene names | EFCAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.010700 (rank : 105) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
HNRL1_MOUSE
|
||||||
| NC score | 0.010630 (rank : 106) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
SSBP2_MOUSE
|
||||||
| NC score | 0.010493 (rank : 107) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CYZ8, Q99LX9 | Gene names | Ssbp2, Ssdp2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 2 (Sequence-specific single- stranded-DNA-binding protein 2). | |||||
|
SSBP2_HUMAN
|
||||||
| NC score | 0.010474 (rank : 108) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P81877, Q8N2Q2, Q9BWW6, Q9Y4T7 | Gene names | SSBP2, SSDP2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 2 (Sequence-specific single- stranded-DNA-binding protein 2). | |||||
|
JAD1C_HUMAN
|
||||||
| NC score | 0.010434 (rank : 109) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.010358 (rank : 110) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
PLS3_MOUSE
|
||||||
| NC score | 0.009834 (rank : 111) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | 0.009793 (rank : 112) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
EMID2_HUMAN
|
||||||
| NC score | 0.009704 (rank : 113) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96A83 | Gene names | EMID2, COL26A1, EMU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.009697 (rank : 114) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CCD1A_HUMAN
|
||||||
| NC score | 0.009443 (rank : 115) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.009349 (rank : 116) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.009280 (rank : 117) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
HNF3G_HUMAN
|
||||||
| NC score | 0.009199 (rank : 118) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55318, Q9UMW9 | Gene names | FOXA3, HNF3G, TCF3G | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-gamma (HNF-3G) (Forkhead box protein A3) (Fork head-related protein FKH H3). | |||||
|
JIP4_MOUSE
|
||||||
| NC score | 0.009005 (rank : 119) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.008942 (rank : 120) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.008865 (rank : 121) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
PDLI5_HUMAN
|
||||||
| NC score | 0.008750 (rank : 122) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.008601 (rank : 123) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.008119 (rank : 124) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.008103 (rank : 125) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.008032 (rank : 126) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.008018 (rank : 127) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
TISD_HUMAN
|
||||||
| NC score | 0.007913 (rank : 128) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
PXK_MOUSE
|
||||||
| NC score | 0.007867 (rank : 129) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.007402 (rank : 130) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.006813 (rank : 131) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
K1196_HUMAN
|
||||||
| NC score | 0.006716 (rank : 132) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
CO1A2_MOUSE
|
||||||
| NC score | 0.006635 (rank : 133) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.006494 (rank : 134) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
FOXF2_HUMAN
|
||||||
| NC score | 0.006209 (rank : 135) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12947, Q9UQ85 | Gene names | FOXF2, FKHL6, FREAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein F2 (Forkhead-related protein FKHL6) (Forkhead- related transcription factor 2) (FREAC-2) (Forkhead-related activator 2). | |||||
|
FBLI1_HUMAN
|
||||||
| NC score | 0.006133 (rank : 136) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WUP2, Q5VVE0, Q5VVE1, Q8IX23, Q96T00, Q9UFD6 | Gene names | FBLIM1, FBLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-binding LIM protein 1 (Filamin-binding LIM protein 1) (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin). | |||||
|
OTX1_HUMAN
|
||||||
| NC score | 0.006128 (rank : 137) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P32242, Q53TG6 | Gene names | OTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
KCAB3_HUMAN
|
||||||
| NC score | 0.006088 (rank : 138) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43448 | Gene names | KCNAB3, KCNA3B | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-gated potassium channel subunit beta-3 (K(+) channel subunit beta-3) (Kv-beta-3). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.005944 (rank : 139) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.005824 (rank : 140) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
SCMH1_HUMAN
|
||||||
| NC score | 0.005824 (rank : 141) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96GD3, Q5VT76, Q6IAJ4, Q8WU48, Q9UKM5, Q9UKM6 | Gene names | SCMH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
ANR47_HUMAN
|
||||||
| NC score | 0.005685 (rank : 142) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NY19, Q6NZI1, Q6ZQR3, Q8IUV2 | Gene names | ANKRD47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.005606 (rank : 143) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.005541 (rank : 144) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
NKX61_MOUSE
|
||||||
| NC score | 0.005297 (rank : 145) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.004926 (rank : 146) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.004682 (rank : 147) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.004547 (rank : 148) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
CXA3_HUMAN
|
||||||
| NC score | 0.004307 (rank : 149) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6H8, Q9H537 | Gene names | GJA3 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-3 protein (Connexin-46) (Cx46). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.004267 (rank : 150) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.003829 (rank : 151) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
BTBD2_HUMAN
|
||||||
| NC score | 0.003795 (rank : 152) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BX70, O60418, O75248, Q7Z5W0, Q96SX8, Q9NPS1, Q9NX81 | Gene names | BTBD2 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 2. | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.003728 (rank : 153) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.003505 (rank : 154) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.003426 (rank : 155) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
HXA3_MOUSE
|
||||||
| NC score | 0.003341 (rank : 156) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.002862 (rank : 157) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
OST4_HUMAN
|
||||||
| NC score | 0.002769 (rank : 158) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y661, Q5QI42, Q8NDC2 | Gene names | HS3ST4, 3OST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 4 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4) (Heparan sulfate 3-O-sulfotransferase 4) (h3-OST-4). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.002501 (rank : 159) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
ZN219_HUMAN
|
||||||
| NC score | 0.002202 (rank : 160) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.000539 (rank : 161) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
ACM4_MOUSE
|
||||||
| NC score | 0.000476 (rank : 162) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32211, Q64056 | Gene names | Chrm4, Chrm-4 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M4 (Mm4 mAChR). | |||||
|
MTF1_HUMAN
|
||||||
| NC score | 0.000349 (rank : 163) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
STK39_MOUSE
|
||||||
| NC score | 0.000175 (rank : 164) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |

|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
M3K12_MOUSE
|
||||||
| NC score | -0.000563 (rank : 165) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
CAMKV_MOUSE
|
||||||
| NC score | -0.000751 (rank : 166) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3UHL1, Q8VD20 | Gene names | Camkv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
ACM4_HUMAN
|
||||||
| NC score | -0.000939 (rank : 167) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08173 | Gene names | CHRM4 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M4. | |||||
|
M3K11_HUMAN
|
||||||
| NC score | -0.000997 (rank : 168) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | -0.000999 (rank : 169) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
MK07_MOUSE
|
||||||
| NC score | -0.001818 (rank : 170) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||