Please be patient as the page loads
|
CHSS3_HUMAN
|
||||||
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CHSS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985423 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984780 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CGAT1_HUMAN
|
||||||
| θ value | 4.11246e-53 (rank : 4) | NC score | 0.818296 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_MOUSE
|
||||||
| θ value | 5.37103e-53 (rank : 5) | NC score | 0.820093 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_MOUSE
|
||||||
| θ value | 1.23823e-49 (rank : 6) | NC score | 0.818030 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_HUMAN
|
||||||
| θ value | 2.75849e-49 (rank : 7) | NC score | 0.818138 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHGUT_HUMAN
|
||||||
| θ value | 1.28434e-38 (rank : 8) | NC score | 0.718281 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
CHSS2_MOUSE
|
||||||
| θ value | 4.13158e-37 (rank : 9) | NC score | 0.723099 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
CHSS2_HUMAN
|
||||||
| θ value | 3.27365e-34 (rank : 10) | NC score | 0.719303 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
B4GN3_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 11) | NC score | 0.460866 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN3_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 12) | NC score | 0.466130 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 13) | NC score | 0.470132 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 14) | NC score | 0.471310 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B3GLT_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 15) | NC score | 0.357285 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B3GLT_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 16) | NC score | 0.342602 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
RFNG_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 17) | NC score | 0.245429 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09009 | Gene names | Rfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 18) | NC score | 0.237310 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O09010, Q3U659, Q8K3F1, Q9DC10 | Gene names | Lfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 19) | NC score | 0.223964 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NES3, O00589, Q96C39, Q9UJW5 | Gene names | LFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
MFNG_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 20) | NC score | 0.182262 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09008 | Gene names | Mfng | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GT2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 21) | NC score | 0.168660 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 22) | NC score | 0.167682 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.061669 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.061674 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
B4GT1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.130862 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.142877 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
RFNG_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.219005 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y644, O00588 | Gene names | RFNG | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.020043 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.026919 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
B4GT4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.120792 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.024920 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.034960 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.017563 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.024621 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.016399 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.040082 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.025782 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
GLT12_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.020617 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IXK2, Q5TCF7, Q8NG54, Q96CT9, Q9H771 | Gene names | GALNT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
MFNG_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.161713 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00587, O43730 | Gene names | MFNG | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B3GL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.052087 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75752, Q3Y531, Q6IAI5, Q8NFM8, Q9HA06 | Gene names | B3GALNT1, B3GALT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
CU087_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.057337 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59051 | Gene names | C21orf87 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf87. | |||||
|
IF4B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.036426 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
KCD12_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.024572 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96CX2 | Gene names | KCTD12, C13orf2, KIAA1778, PFET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.018503 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
B4GT4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.116314 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GDPD5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.025925 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q640M6, Q8R0T5, Q8R3N5 | Gene names | Gdpd5, Gde2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycerophosphodiester phosphodiesterase domain-containing protein 5 (EC 3.1.-.-) (Glycerophosphodiester phosphodiesterase 2). | |||||
|
GLT12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.020152 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.014320 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
KCD12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.022489 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6WVG3 | Gene names | Kctd12, Pfet1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.014739 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ZN212_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.000988 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UDV6, Q13396, Q8N664 | Gene names | ZNF212, ZNFC150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 212 (Zinc finger protein C2H2-150). | |||||
|
B4GT7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.091369 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.093599 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
GDPD5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.024486 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WTR4, Q49AQ5, Q6UX76, Q7Z4S0, Q8N781, Q8NCB7, Q8TB77 | Gene names | GDPD5, GDE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycerophosphodiester phosphodiesterase domain-containing protein 5 (EC 3.1.-.-) (Glycerophosphodiester phosphodiesterase 2). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.019754 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.021502 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.009679 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
B3A4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.009222 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96Q91, Q96RM5, Q9BXF2, Q9BXN3 | Gene names | SLC4A9, AE4, SBC5 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 4 (Anion exchanger 4) (Solute carrier family 4 member 9) (Sodium bicarbonate cotransporter 5). | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.022537 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.022755 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.016019 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.018029 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.024059 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
B4GT3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.091715 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.092544 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
SEN34_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.020336 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BSV6, Q9BVT1, Q9H6H5 | Gene names | TSEN34, LENG5, SEN34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (HsSen34) (Leukocyte receptor cluster member 5). | |||||
|
SNX19_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.020179 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92543 | Gene names | SNX19, KIAA0254 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-19. | |||||
|
SON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.015932 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
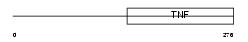
TNFL6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.018034 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48023, Q9BZP9 | Gene names | FASLG, APT1LG1, FASL, TNFSF6 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor ligand superfamily member 6 (Fas antigen ligand) (Fas ligand) (CD178 antigen) (CD95L protein) (Apoptosis antigen ligand) (APTL) [Contains: Tumor necrosis factor ligand superfamily member 6, membrane form; Tumor necrosis factor ligand superfamily member 6, soluble form]. | |||||
|
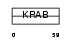
ZN133_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | -0.001986 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52736, Q9BUV2, Q9H443 | Gene names | ZNF133 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 133. | |||||
|
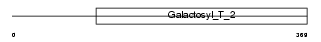
B4GT6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.088635 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.088404 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
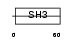
CASL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.009439 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |
|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.023145 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
GALT4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.017550 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N4A0, O00208 | Gene names | GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GALT4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.017876 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08832, Q3U681 | Gene names | Galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.004631 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
B3GL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.033650 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q920V1, O54906, Q91V72, Q920V0, Q9CTE5 | Gene names | b3galnt1, B3galt3, B3gt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
B3GT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.020029 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5Z6 | Gene names | B3GALT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,3-GalTase 1) (Beta3GalT1) (UDP-galactose:beta-N-acetyl-glucosamine-beta-1,3- galactosyltransferase 1). | |||||
|
B3GT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.020029 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54904, Q91V52 | Gene names | B3galt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,3-GalTase 1) (Beta3GalT1) (UDP-galactose:beta-N-acetyl-glucosamine-beta-1,3- galactosyltransferase 1) (UDP-Gal:betaGlcNAc beta 1,3- galactosyltransferase-I). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.015375 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
ANGL6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.003561 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI99, Q9BZZ0 | Gene names | ANGPTL6, AGF, ARP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 6 precursor (Angiopoietin-like 6) (Angiopoietin-related growth factor) (Angiopoietin-related protein 5). | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.022505 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.017582 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
CSTF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.007178 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
GALT6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.018292 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
RPKL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.000156 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R2S1, Q8R1D4 | Gene names | Rps6kl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase-like 1 (EC 2.7.11.1). | |||||
|
ZN185_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.014578 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
|
ZN282_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | -0.001543 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UDV7, O43691, Q6DKK0 | Gene names | ZNF282, HUB1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 282 (HTLV-I U5RE-binding protein 1) (HUB-1). | |||||
|
B3GN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.016171 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NY97, Q9NQQ9, Q9NQR0, Q9NUT9 | Gene names | B3GNT1, B3GALT7 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (EC 2.4.1.-) (Beta3Gn-T1) (BGnT-1) (Beta-1,3-galactosyltransferase 7) (Beta-1,3-GalTase 7) (Beta3Gal-T7) (b3Gal-T7) (UDP-galactose:beta-N- acetylglucosamine beta-1,3-galactosyltransferase 7) (UDP-Gal:beta- GlcNAc beta-1,3-galactosyltransferase 7) (Beta-3-Gx-T7). | |||||
|
CENPQ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.009894 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CPQ5, Q8C2G4, Q8VEF8, Q9CX70 | Gene names | Cenpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
CO2A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.021481 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |
|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.013967 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.013304 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.012145 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.006946 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
TMUB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.011096 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BVT8 | Gene names | TMUB1, C7orf21 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1 (Ubiquitin-like protein SB144). | |||||
|
B4GT5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.083258 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.083652 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CHSS1_HUMAN
|
||||||
| NC score | 0.985423 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.984780 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CGAT1_MOUSE
|
||||||
| NC score | 0.820093 (rank : 4) | θ value | 5.37103e-53 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_HUMAN
|
||||||
| NC score | 0.818296 (rank : 5) | θ value | 4.11246e-53 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| NC score | 0.818138 (rank : 6) | θ value | 2.75849e-49 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| NC score | 0.818030 (rank : 7) | θ value | 1.23823e-49 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHSS2_MOUSE
|
||||||
| NC score | 0.723099 (rank : 8) | θ value | 4.13158e-37 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
CHSS2_HUMAN
|
||||||
| NC score | 0.719303 (rank : 9) | θ value | 3.27365e-34 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHGUT_HUMAN
|
||||||
| NC score | 0.718281 (rank : 10) | θ value | 1.28434e-38 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.471310 (rank : 11) | θ value | 2.79066e-09 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.470132 (rank : 12) | θ value | 2.13673e-09 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN3_MOUSE
|
||||||
| NC score | 0.466130 (rank : 13) | θ value | 1.25267e-09 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN3_HUMAN
|
||||||
| NC score | 0.460866 (rank : 14) | θ value | 6.64225e-11 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B3GLT_MOUSE
|
||||||
| NC score | 0.357285 (rank : 15) | θ value | 4.0297e-08 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B3GLT_HUMAN
|
||||||
| NC score | 0.342602 (rank : 16) | θ value | 3.41135e-07 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
RFNG_MOUSE
|
||||||
| NC score | 0.245429 (rank : 17) | θ value | 4.1701e-05 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09009 | Gene names | Rfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_MOUSE
|
||||||
| NC score | 0.237310 (rank : 18) | θ value | 0.000121331 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O09010, Q3U659, Q8K3F1, Q9DC10 | Gene names | Lfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_HUMAN
|
||||||
| NC score | 0.223964 (rank : 19) | θ value | 0.00102713 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NES3, O00589, Q96C39, Q9UJW5 | Gene names | LFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
RFNG_HUMAN
|
||||||
| NC score | 0.219005 (rank : 20) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y644, O00588 | Gene names | RFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
MFNG_MOUSE
|
||||||
| NC score | 0.182262 (rank : 21) | θ value | 0.00298849 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09008 | Gene names | Mfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GT2_HUMAN
|
||||||
| NC score | 0.168660 (rank : 22) | θ value | 0.00869519 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| NC score | 0.167682 (rank : 23) | θ value | 0.00869519 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
MFNG_HUMAN
|
||||||
| NC score | 0.161713 (rank : 24) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00587, O43730 | Gene names | MFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GT1_MOUSE
|
||||||
| NC score | 0.142877 (rank : 25) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.130862 (rank : 26) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_MOUSE
|
||||||
| NC score | 0.120792 (rank : 27) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_HUMAN
|
||||||
| NC score | 0.116314 (rank : 28) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT7_MOUSE
|
||||||
| NC score | 0.093599 (rank : 29) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT3_MOUSE
|
||||||
| NC score | 0.092544 (rank : 30) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_HUMAN
|
||||||
| NC score | 0.091715 (rank : 31) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT7_HUMAN
|
||||||
| NC score | 0.091369 (rank : 32) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT6_HUMAN
|
||||||
| NC score | 0.088635 (rank : 33) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| NC score | 0.088404 (rank : 34) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT5_MOUSE
|
||||||
| NC score | 0.083652 (rank : 35) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_HUMAN
|
||||||
| NC score | 0.083258 (rank : 36) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.061674 (rank : 37) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.061669 (rank : 38) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
CU087_HUMAN
|
||||||
| NC score | 0.057337 (rank : 39) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59051 | Gene names | C21orf87 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf87. | |||||
|
B3GL1_HUMAN
|
||||||
| NC score | 0.052087 (rank : 40) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75752, Q3Y531, Q6IAI5, Q8NFM8, Q9HA06 | Gene names | B3GALNT1, B3GALT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.040082 (rank : 41) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
IF4B_HUMAN
|
||||||
| NC score | 0.036426 (rank : 42) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.034960 (rank : 43) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
B3GL1_MOUSE
|
||||||
| NC score | 0.033650 (rank : 44) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q920V1, O54906, Q91V72, Q920V0, Q9CTE5 | Gene names | b3galnt1, B3galt3, B3gt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.026919 (rank : 45) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
GDPD5_MOUSE
|
||||||
| NC score | 0.025925 (rank : 46) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q640M6, Q8R0T5, Q8R3N5 | Gene names | Gdpd5, Gde2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycerophosphodiester phosphodiesterase domain-containing protein 5 (EC 3.1.-.-) (Glycerophosphodiester phosphodiesterase 2). | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.025782 (rank : 47) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.024920 (rank : 48) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.024621 (rank : 49) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
KCD12_HUMAN
|
||||||
| NC score | 0.024572 (rank : 50) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96CX2 | Gene names | KCTD12, C13orf2, KIAA1778, PFET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
GDPD5_HUMAN
|
||||||
| NC score | 0.024486 (rank : 51) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WTR4, Q49AQ5, Q6UX76, Q7Z4S0, Q8N781, Q8NCB7, Q8TB77 | Gene names | GDPD5, GDE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycerophosphodiester phosphodiesterase domain-containing protein 5 (EC 3.1.-.-) (Glycerophosphodiester phosphodiesterase 2). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.024059 (rank : 52) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.023145 (rank : 53) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
COBA1_MOUSE
|
||||||
| NC score | 0.022755 (rank : 54) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.022537 (rank : 55) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.022505 (rank : 56) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
KCD12_MOUSE
|
||||||
| NC score | 0.022489 (rank : 57) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6WVG3 | Gene names | Kctd12, Pfet1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.021502 (rank : 58) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
CO2A1_HUMAN
|
||||||
| NC score | 0.021481 (rank : 59) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
GLT12_HUMAN
|
||||||
| NC score | 0.020617 (rank : 60) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IXK2, Q5TCF7, Q8NG54, Q96CT9, Q9H771 | Gene names | GALNT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
SEN34_HUMAN
|
||||||
| NC score | 0.020336 (rank : 61) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BSV6, Q9BVT1, Q9H6H5 | Gene names | TSEN34, LENG5, SEN34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (HsSen34) (Leukocyte receptor cluster member 5). | |||||
|
SNX19_HUMAN
|
||||||
| NC score | 0.020179 (rank : 62) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92543 | Gene names | SNX19, KIAA0254 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-19. | |||||
|
GLT12_MOUSE
|
||||||
| NC score | 0.020152 (rank : 63) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.020043 (rank : 64) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
B3GT1_HUMAN
|
||||||
| NC score | 0.020029 (rank : 65) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5Z6 | Gene names | B3GALT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,3-GalTase 1) (Beta3GalT1) (UDP-galactose:beta-N-acetyl-glucosamine-beta-1,3- galactosyltransferase 1). | |||||
|
B3GT1_MOUSE
|
||||||
| NC score | 0.020029 (rank : 66) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54904, Q91V52 | Gene names | B3galt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,3-GalTase 1) (Beta3GalT1) (UDP-galactose:beta-N-acetyl-glucosamine-beta-1,3- galactosyltransferase 1) (UDP-Gal:betaGlcNAc beta 1,3- galactosyltransferase-I). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.019754 (rank : 67) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.018503 (rank : 68) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
GALT6_HUMAN
|
||||||
| NC score | 0.018292 (rank : 69) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
TNFL6_HUMAN
|
||||||
| NC score | 0.018034 (rank : 70) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48023, Q9BZP9 | Gene names | FASLG, APT1LG1, FASL, TNFSF6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor ligand superfamily member 6 (Fas antigen ligand) (Fas ligand) (CD178 antigen) (CD95L protein) (Apoptosis antigen ligand) (APTL) [Contains: Tumor necrosis factor ligand superfamily member 6, membrane form; Tumor necrosis factor ligand superfamily member 6, soluble form]. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.018029 (rank : 71) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
GALT4_MOUSE
|
||||||
| NC score | 0.017876 (rank : 72) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08832, Q3U681 | Gene names | Galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.017582 (rank : 73) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.017563 (rank : 74) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
GALT4_HUMAN
|
||||||
| NC score | 0.017550 (rank : 75) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N4A0, O00208 | Gene names | GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.016399 (rank : 76) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
B3GN1_HUMAN
|
||||||
| NC score | 0.016171 (rank : 77) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NY97, Q9NQQ9, Q9NQR0, Q9NUT9 | Gene names | B3GNT1, B3GALT7 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (EC 2.4.1.-) (Beta3Gn-T1) (BGnT-1) (Beta-1,3-galactosyltransferase 7) (Beta-1,3-GalTase 7) (Beta3Gal-T7) (b3Gal-T7) (UDP-galactose:beta-N- acetylglucosamine beta-1,3-galactosyltransferase 7) (UDP-Gal:beta- GlcNAc beta-1,3-galactosyltransferase 7) (Beta-3-Gx-T7). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.016019 (rank : 78) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.015932 (rank : 79) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.015375 (rank : 80) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.014739 (rank : 81) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ZN185_HUMAN
|
||||||
| NC score | 0.014578 (rank : 82) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.014320 (rank : 83) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.013967 (rank : 84) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.013304 (rank : 85) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.012145 (rank : 86) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
TMUB1_HUMAN
|
||||||
| NC score | 0.011096 (rank : 87) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BVT8 | Gene names | TMUB1, C7orf21 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1 (Ubiquitin-like protein SB144). | |||||
|
CENPQ_MOUSE
|
||||||
| NC score | 0.009894 (rank : 88) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CPQ5, Q8C2G4, Q8VEF8, Q9CX70 | Gene names | Cenpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.009679 (rank : 89) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
CASL_MOUSE
|
||||||
| NC score | 0.009439 (rank : 90) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
B3A4_HUMAN
|
||||||
| NC score | 0.009222 (rank : 91) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96Q91, Q96RM5, Q9BXF2, Q9BXN3 | Gene names | SLC4A9, AE4, SBC5 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 4 (Anion exchanger 4) (Solute carrier family 4 member 9) (Sodium bicarbonate cotransporter 5). | |||||
|
CSTF2_MOUSE
|
||||||
| NC score | 0.007178 (rank : 92) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.006946 (rank : 93) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.004631 (rank : 94) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ANGL6_HUMAN
|
||||||
| NC score | 0.003561 (rank : 95) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI99, Q9BZZ0 | Gene names | ANGPTL6, AGF, ARP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 6 precursor (Angiopoietin-like 6) (Angiopoietin-related growth factor) (Angiopoietin-related protein 5). | |||||
|
ZN212_HUMAN
|
||||||
| NC score | 0.000988 (rank : 96) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UDV6, Q13396, Q8N664 | Gene names | ZNF212, ZNFC150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 212 (Zinc finger protein C2H2-150). | |||||
|
RPKL1_MOUSE
|
||||||
| NC score | 0.000156 (rank : 97) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R2S1, Q8R1D4 | Gene names | Rps6kl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase-like 1 (EC 2.7.11.1). | |||||
|
ZN282_HUMAN
|
||||||
| NC score | -0.001543 (rank : 98) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UDV7, O43691, Q6DKK0 | Gene names | ZNF282, HUB1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 282 (HTLV-I U5RE-binding protein 1) (HUB-1). | |||||
|
ZN133_HUMAN
|
||||||
| NC score | -0.001986 (rank : 99) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52736, Q9BUV2, Q9H443 | Gene names | ZNF133 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 133. | |||||