Please be patient as the page loads
|
B4GT1_MOUSE
|
||||||
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
B4GT1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.982690 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_HUMAN
|
||||||
| θ value | 6.24247e-110 (rank : 3) | NC score | 0.986316 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| θ value | 2.6227e-108 (rank : 4) | NC score | 0.986912 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_MOUSE
|
||||||
| θ value | 2.30828e-88 (rank : 5) | NC score | 0.977970 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_HUMAN
|
||||||
| θ value | 5.14231e-88 (rank : 6) | NC score | 0.976128 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_MOUSE
|
||||||
| θ value | 2.24094e-75 (rank : 7) | NC score | 0.974915 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_HUMAN
|
||||||
| θ value | 1.11217e-74 (rank : 8) | NC score | 0.972979 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT5_MOUSE
|
||||||
| θ value | 2.4834e-66 (rank : 9) | NC score | 0.949508 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_HUMAN
|
||||||
| θ value | 4.68348e-65 (rank : 10) | NC score | 0.949209 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT6_MOUSE
|
||||||
| θ value | 3.14151e-61 (rank : 11) | NC score | 0.945638 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_HUMAN
|
||||||
| θ value | 4.10295e-61 (rank : 12) | NC score | 0.946570 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT7_MOUSE
|
||||||
| θ value | 5.77852e-31 (rank : 13) | NC score | 0.886019 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT7_HUMAN
|
||||||
| θ value | 1.4233e-29 (rank : 14) | NC score | 0.878659 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
CHSS1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.169006 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.162421 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.142877 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.016660 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
RASF7_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.059856 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DD19 | Gene names | Rassf7 | |||
|
Domain Architecture |
|
|||||
| Description | Ras association domain-containing protein 7. | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.018614 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CGAT1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.106368 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.101490 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.103466 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.104117 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
LBN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.022249 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86UK5, Q86YT3, Q86YT4, Q8NG49 | Gene names | EVC2, LBN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.020468 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
CO4A1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.017221 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.001092 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.011266 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
WNT3A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.011702 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27467 | Gene names | Wnt3a, Wnt-3a | |||
|
Domain Architecture |
|
|||||
| Description | Protein Wnt-3a precursor. | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.019226 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.017336 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
ULK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.000692 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
WNT3A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.010658 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56704, Q969P2 | Gene names | WNT3A | |||
|
Domain Architecture |
|
|||||
| Description | Protein Wnt-3a precursor. | |||||
|
XKR5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.012839 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UX68, Q5GH74 | Gene names | XKR5, XRG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 5. | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.010672 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
FOXK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.004274 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.009919 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
BARH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.001623 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63157, Q9JK26 | Gene names | Barhl1, Bhx1, Mbh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BarH-like 1 homeobox protein (Bar-class homeodomain protein MBH2) (BarH-related homeobox protein 1). | |||||
|
ULK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.000473 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
ANKS6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.001505 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.003406 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.009761 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
DOK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.007134 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99704, O43204, Q9UHG6 | Gene names | DOK1 | |||
|
Domain Architecture |
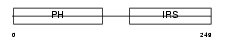
|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)) (pp62). | |||||
|
I17RA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.008604 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96F46, O43844 | Gene names | IL17RA, IL17R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-17 receptor A precursor (IL-17 receptor). | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.012685 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.007985 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
ASTL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.005914 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
PLIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.007062 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60240, Q8N5Y6 | Gene names | PLIN | |||
|
Domain Architecture |

|
|||||
| Description | Perilipin (PERI) (Lipid droplet-associated protein). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.005667 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.077835 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.067415 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GT1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| NC score | 0.986912 (rank : 2) | θ value | 2.6227e-108 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_HUMAN
|
||||||
| NC score | 0.986316 (rank : 3) | θ value | 6.24247e-110 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.982690 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_MOUSE
|
||||||
| NC score | 0.977970 (rank : 5) | θ value | 2.30828e-88 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_HUMAN
|
||||||
| NC score | 0.976128 (rank : 6) | θ value | 5.14231e-88 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_MOUSE
|
||||||
| NC score | 0.974915 (rank : 7) | θ value | 2.24094e-75 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_HUMAN
|
||||||
| NC score | 0.972979 (rank : 8) | θ value | 1.11217e-74 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT5_MOUSE
|
||||||
| NC score | 0.949508 (rank : 9) | θ value | 2.4834e-66 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_HUMAN
|
||||||
| NC score | 0.949209 (rank : 10) | θ value | 4.68348e-65 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT6_HUMAN
|
||||||
| NC score | 0.946570 (rank : 11) | θ value | 4.10295e-61 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| NC score | 0.945638 (rank : 12) | θ value | 3.14151e-61 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT7_MOUSE
|
||||||
| NC score | 0.886019 (rank : 13) | θ value | 5.77852e-31 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT7_HUMAN
|
||||||
| NC score | 0.878659 (rank : 14) | θ value | 1.4233e-29 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
CHSS1_HUMAN
|
||||||
| NC score | 0.169006 (rank : 15) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.162421 (rank : 16) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.142877 (rank : 17) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CGAT1_MOUSE
|
||||||
| NC score | 0.106368 (rank : 18) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_MOUSE
|
||||||
| NC score | 0.104117 (rank : 19) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT1_HUMAN
|
||||||
| NC score | 0.103466 (rank : 20) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| NC score | 0.101490 (rank : 21) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.077835 (rank : 22) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.067415 (rank : 23) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
RASF7_MOUSE
|
||||||
| NC score | 0.059856 (rank : 24) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DD19 | Gene names | Rassf7 | |||
|
Domain Architecture |

|
|||||
| Description | Ras association domain-containing protein 7. | |||||
|
LBN_HUMAN
|
||||||
| NC score | 0.022249 (rank : 25) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86UK5, Q86YT3, Q86YT4, Q8NG49 | Gene names | EVC2, LBN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.020468 (rank : 26) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.019226 (rank : 27) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.018614 (rank : 28) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.017336 (rank : 29) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
CO4A1_MOUSE
|
||||||
| NC score | 0.017221 (rank : 30) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.016660 (rank : 31) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
XKR5_HUMAN
|
||||||
| NC score | 0.012839 (rank : 32) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UX68, Q5GH74 | Gene names | XKR5, XRG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 5. | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.012685 (rank : 33) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
WNT3A_MOUSE
|
||||||
| NC score | 0.011702 (rank : 34) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27467 | Gene names | Wnt3a, Wnt-3a | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-3a precursor. | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.011266 (rank : 35) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.010672 (rank : 36) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
WNT3A_HUMAN
|
||||||
| NC score | 0.010658 (rank : 37) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56704, Q969P2 | Gene names | WNT3A | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-3a precursor. | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.009919 (rank : 38) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.009761 (rank : 39) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
I17RA_HUMAN
|
||||||
| NC score | 0.008604 (rank : 40) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96F46, O43844 | Gene names | IL17RA, IL17R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-17 receptor A precursor (IL-17 receptor). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.007985 (rank : 41) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
DOK1_HUMAN
|
||||||
| NC score | 0.007134 (rank : 42) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99704, O43204, Q9UHG6 | Gene names | DOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)) (pp62). | |||||
|
PLIN_HUMAN
|
||||||
| NC score | 0.007062 (rank : 43) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60240, Q8N5Y6 | Gene names | PLIN | |||
|
Domain Architecture |

|
|||||
| Description | Perilipin (PERI) (Lipid droplet-associated protein). | |||||
|
ASTL_HUMAN
|
||||||
| NC score | 0.005914 (rank : 44) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.005667 (rank : 45) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
FOXK1_MOUSE
|
||||||
| NC score | 0.004274 (rank : 46) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.003406 (rank : 47) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
BARH1_MOUSE
|
||||||
| NC score | 0.001623 (rank : 48) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63157, Q9JK26 | Gene names | Barhl1, Bhx1, Mbh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BarH-like 1 homeobox protein (Bar-class homeodomain protein MBH2) (BarH-related homeobox protein 1). | |||||
|
ANKS6_HUMAN
|
||||||
| NC score | 0.001505 (rank : 49) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.001092 (rank : 50) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ULK2_MOUSE
|
||||||
| NC score | 0.000692 (rank : 51) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
ULK2_HUMAN
|
||||||
| NC score | 0.000473 (rank : 52) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||