Please be patient as the page loads
|
CN004_MOUSE
|
||||||
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CN004_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.977566 (rank : 2) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
CN004_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 3) | NC score | 0.166725 (rank : 3) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
SEM4G_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 4) | NC score | 0.052846 (rank : 33) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 5) | NC score | 0.082933 (rank : 6) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.065493 (rank : 14) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.065946 (rank : 13) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.039891 (rank : 53) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.056629 (rank : 24) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.069764 (rank : 11) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.039392 (rank : 55) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.039446 (rank : 54) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
BAI1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.026760 (rank : 79) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
TRI47_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.070697 (rank : 10) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96LD4, Q96GU5, Q9BRN7 | Gene names | TRIM47, GOA, RNF100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47 (Gene overexpressed in astrocytoma protein) (RING finger protein 100). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.046099 (rank : 45) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
EPHA6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.014606 (rank : 105) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UF33 | Gene names | EPHA6, EHK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephrin type-A receptor 6 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-2) (EPH homology kinase 2). | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.064435 (rank : 16) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.072399 (rank : 9) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
ZIMP7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.041251 (rank : 50) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.065087 (rank : 15) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ZN516_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.011013 (rank : 110) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
LRC56_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.054884 (rank : 26) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.061446 (rank : 18) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.059613 (rank : 22) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RFWD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.046866 (rank : 42) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
RFWD2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.047619 (rank : 41) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.009089 (rank : 112) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.084421 (rank : 5) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.080887 (rank : 7) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
TLX3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.013500 (rank : 106) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43711, Q96AD3 | Gene names | TLX3, HOX11L2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell leukemia homeobox protein 3 (Homeobox protein Hox-11L2). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.035048 (rank : 64) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.045109 (rank : 46) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.035737 (rank : 63) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
AMOL2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.023032 (rank : 91) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
B4GT1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.025107 (rank : 84) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.034661 (rank : 68) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.023286 (rank : 90) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
HAIR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.060108 (rank : 20) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
TRI47_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.053628 (rank : 30) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C0E3, Q6P249, Q811J7, Q8BVZ8, Q8R1K0, Q8R3Y1 | Gene names | Trim47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.088654 (rank : 4) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DCX_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.037753 (rank : 56) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
DPOLN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.041437 (rank : 49) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z5Q5, Q4TTW4, Q6ZNF4 | Gene names | POLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase nu (EC 2.7.7.7). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.037621 (rank : 57) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.053919 (rank : 29) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
TRIM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.037514 (rank : 58) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJV3, Q5JYF5, Q8WWK1, Q9UJR9 | Gene names | MID2, FXY2, RNF60, TRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1) (Midin-2) (RING finger protein 60). | |||||
|
TRIM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.037169 (rank : 60) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QUS6 | Gene names | Mid2, Fxy2, Trim1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.010299 (rank : 111) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ANR38_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.013156 (rank : 108) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T7N3, Q6P9A0, Q86T71, Q86VE6, Q8NAX3 | Gene names | ANKRD38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
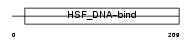
HSF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.041743 (rank : 48) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
MK15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.007948 (rank : 117) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.033184 (rank : 71) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
ZN500_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.000550 (rank : 120) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60304, Q96CQ8, Q9BTG0 | Gene names | ZNF500, KIAA0557 | |||
|
Domain Architecture |
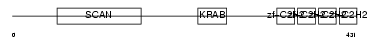
|
|||||
| Description | Zinc finger protein 500. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.044436 (rank : 47) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.025959 (rank : 81) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
ONEC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.033040 (rank : 72) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95948 | Gene names | ONECUT2 | |||
|
Domain Architecture |

|
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
ONEC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.032806 (rank : 73) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6XBJ3 | Gene names | Onecut2, Oc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
PANK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.017956 (rank : 99) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |
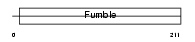
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.033582 (rank : 70) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.037380 (rank : 59) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PP2CG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.018424 (rank : 96) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.029680 (rank : 75) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TCF19_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.051916 (rank : 34) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
TRIP6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.035040 (rank : 65) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.013402 (rank : 107) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CSRP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.020826 (rank : 93) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97314, Q9CZG5 | Gene names | Csrp2, Dlp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Double LIM protein 1) (DLP-1). | |||||
|
DAB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.027636 (rank : 78) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |
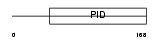
|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
HSF4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.036517 (rank : 61) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0L1, Q9R0L2 | Gene names | Hsf4 | |||
|
Domain Architecture |
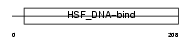
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (mHSF4). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.025906 (rank : 82) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.046635 (rank : 43) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
OAS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.017279 (rank : 101) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.024299 (rank : 86) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.030106 (rank : 74) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.046106 (rank : 44) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SENP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.016494 (rank : 103) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H4L4, Q96PS4, Q9Y3W9 | Gene names | SENP3, SSP3, SUSP3 | |||
|
Domain Architecture |
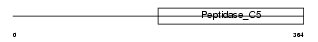
|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3). | |||||
|
TRI46_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.034991 (rank : 67) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z4K8, Q6NT17, Q6NT41, Q6ZRL7, Q9H5P2 | Gene names | TRIM46, TRIFIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 46 (Tripartite, fibronectin type- III and C-terminal SPRY motif protein). | |||||
|
TRI46_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.035037 (rank : 66) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TNM2, Q60717, Q6P1I9 | Gene names | Trim46, Trific | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 46 (Tripartite, fibronectin type- III and C-terminal SPRY motif protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.018376 (rank : 97) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.015923 (rank : 104) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.036445 (rank : 62) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
LAP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.005508 (rank : 119) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.026754 (rank : 80) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.040755 (rank : 52) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.048108 (rank : 40) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.022749 (rank : 92) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
SERA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.027835 (rank : 76) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61753, Q75SV9 | Gene names | Phgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH) (A10). | |||||
|
ARRD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.017625 (rank : 100) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
BEGIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.025271 (rank : 83) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.050383 (rank : 39) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.023947 (rank : 87) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CO002_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.040877 (rank : 51) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.020570 (rank : 94) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.008080 (rank : 116) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
GNAS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.020106 (rank : 95) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
GP124_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.008525 (rank : 115) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.011628 (rank : 109) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
LAMB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.006336 (rank : 118) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.018340 (rank : 98) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.034376 (rank : 69) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.016982 (rank : 102) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
SERA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.025067 (rank : 85) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43175, Q5SZU3, Q9BQ01 | Gene names | PHGDH, PGDH3 | |||
|
Domain Architecture |
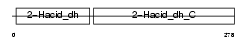
|
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.027813 (rank : 77) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
TRI59_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.023941 (rank : 88) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IWR1 | Gene names | TRIM59, RNF104, TRIM57, TSBF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (Tumor suppressor TSBF-1) (RING finger protein 104). | |||||
|
TRI59_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.023917 (rank : 89) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q922Y2, Q9CSP2, Q9CUD5, Q9D740 | Gene names | Trim59, Mrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (RING finger protein 1). | |||||
|
WNT5A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.008860 (rank : 114) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41221 | Gene names | WNT5A | |||
|
Domain Architecture |
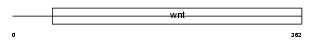
|
|||||
| Description | Protein Wnt-5a precursor. | |||||
|
WNT5A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.008884 (rank : 113) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22725 | Gene names | Wnt5a, Wnt-5a | |||
|
Domain Architecture |
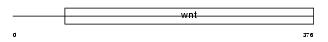
|
|||||
| Description | Protein Wnt-5a precursor. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.053077 (rank : 32) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.050690 (rank : 37) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.054759 (rank : 27) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.050533 (rank : 38) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.060014 (rank : 21) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.060219 (rank : 19) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.055940 (rank : 25) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.073647 (rank : 8) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.067635 (rank : 12) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051096 (rank : 36) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.051343 (rank : 35) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.062071 (rank : 17) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.056716 (rank : 23) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.053549 (rank : 31) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.054174 (rank : 28) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
CN004_HUMAN
|
||||||
| NC score | 0.977566 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.166725 (rank : 3) | θ value | 0.00298849 (rank : 3) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.088654 (rank : 4) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.084421 (rank : 5) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.082933 (rank : 6) | θ value | 0.0193708 (rank : 5) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.080887 (rank : 7) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.073647 (rank : 8) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.072399 (rank : 9) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
TRI47_HUMAN
|
||||||
| NC score | 0.070697 (rank : 10) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96LD4, Q96GU5, Q9BRN7 | Gene names | TRIM47, GOA, RNF100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47 (Gene overexpressed in astrocytoma protein) (RING finger protein 100). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.069764 (rank : 11) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.067635 (rank : 12) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.065946 (rank : 13) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.065493 (rank : 14) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.065087 (rank : 15) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.064435 (rank : 16) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.062071 (rank : 17) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.061446 (rank : 18) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.060219 (rank : 19) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
HAIR_MOUSE
|
||||||
| NC score | 0.060108 (rank : 20) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.060014 (rank : 21) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.059613 (rank : 22) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.056716 (rank : 23) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.056629 (rank : 24) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.055940 (rank : 25) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
LRC56_HUMAN
|
||||||
| NC score | 0.054884 (rank : 26) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.054759 (rank : 27) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.054174 (rank : 28) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.053919 (rank : 29) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
TRI47_MOUSE
|
||||||
| NC score | 0.053628 (rank : 30) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C0E3, Q6P249, Q811J7, Q8BVZ8, Q8R1K0, Q8R3Y1 | Gene names | Trim47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.053549 (rank : 31) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.053077 (rank : 32) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
SEM4G_MOUSE
|
||||||
| NC score | 0.052846 (rank : 33) | θ value | 0.00869519 (rank : 4) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
TCF19_HUMAN
|
||||||
| NC score | 0.051916 (rank : 34) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.051343 (rank : 35) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.051096 (rank : 36) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.050690 (rank : 37) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.050533 (rank : 38) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.050383 (rank : 39) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.048108 (rank : 40) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RFWD2_MOUSE
|
||||||
| NC score | 0.047619 (rank : 41) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
RFWD2_HUMAN
|
||||||
| NC score | 0.046866 (rank : 42) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.046635 (rank : 43) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.046106 (rank : 44) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.046099 (rank : 45) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.045109 (rank : 46) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.044436 (rank : 47) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HSF4_HUMAN
|
||||||
| NC score | 0.041743 (rank : 48) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
DPOLN_HUMAN
|
||||||
| NC score | 0.041437 (rank : 49) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z5Q5, Q4TTW4, Q6ZNF4 | Gene names | POLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase nu (EC 2.7.7.7). | |||||
|
ZIMP7_HUMAN
|
||||||
| NC score | 0.041251 (rank : 50) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.040877 (rank : 51) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.040755 (rank : 52) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.039891 (rank : 53) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.039446 (rank : 54) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.039392 (rank : 55) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.037753 (rank : 56) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.037621 (rank : 57) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
TRIM1_HUMAN
|
||||||
| NC score | 0.037514 (rank : 58) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJV3, Q5JYF5, Q8WWK1, Q9UJR9 | Gene names | MID2, FXY2, RNF60, TRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1) (Midin-2) (RING finger protein 60). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.037380 (rank : 59) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
TRIM1_MOUSE
|
||||||
| NC score | 0.037169 (rank : 60) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QUS6 | Gene names | Mid2, Fxy2, Trim1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1). | |||||
|
HSF4_MOUSE
|
||||||
| NC score | 0.036517 (rank : 61) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0L1, Q9R0L2 | Gene names | Hsf4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (mHSF4). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.036445 (rank : 62) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.035737 (rank : 63) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.035048 (rank : 64) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
TRIP6_HUMAN
|
||||||
| NC score | 0.035040 (rank : 65) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
TRI46_MOUSE
|
||||||
| NC score | 0.035037 (rank : 66) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TNM2, Q60717, Q6P1I9 | Gene names | Trim46, Trific | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 46 (Tripartite, fibronectin type- III and C-terminal SPRY motif protein). | |||||
|
TRI46_HUMAN
|
||||||
| NC score | 0.034991 (rank : 67) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z4K8, Q6NT17, Q6NT41, Q6ZRL7, Q9H5P2 | Gene names | TRIM46, TRIFIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 46 (Tripartite, fibronectin type- III and C-terminal SPRY motif protein). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.034661 (rank : 68) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.034376 (rank : 69) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.033582 (rank : 70) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.033184 (rank : 71) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
ONEC2_HUMAN
|
||||||
| NC score | 0.033040 (rank : 72) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95948 | Gene names | ONECUT2 | |||
|
Domain Architecture |

|
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
ONEC2_MOUSE
|
||||||
| NC score | 0.032806 (rank : 73) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6XBJ3 | Gene names | Onecut2, Oc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.030106 (rank : 74) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.029680 (rank : 75) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SERA_MOUSE
|
||||||
| NC score | 0.027835 (rank : 76) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61753, Q75SV9 | Gene names | Phgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH) (A10). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.027813 (rank : 77) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
DAB2_MOUSE
|
||||||
| NC score | 0.027636 (rank : 78) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.026760 (rank : 79) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.026754 (rank : 80) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.025959 (rank : 81) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.025906 (rank : 82) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
BEGIN_HUMAN
|
||||||
| NC score | 0.025271 (rank : 83) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.025107 (rank : 84) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
SERA_HUMAN
|
||||||
| NC score | 0.025067 (rank : 85) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43175, Q5SZU3, Q9BQ01 | Gene names | PHGDH, PGDH3 | |||
|
Domain Architecture |

|
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.024299 (rank : 86) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.023947 (rank : 87) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
TRI59_HUMAN
|
||||||
| NC score | 0.023941 (rank : 88) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IWR1 | Gene names | TRIM59, RNF104, TRIM57, TSBF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (Tumor suppressor TSBF-1) (RING finger protein 104). | |||||
|
TRI59_MOUSE
|
||||||
| NC score | 0.023917 (rank : 89) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q922Y2, Q9CSP2, Q9CUD5, Q9D740 | Gene names | Trim59, Mrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (RING finger protein 1). | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.023286 (rank : 90) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
AMOL2_MOUSE
|
||||||
| NC score | 0.023032 (rank : 91) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.022749 (rank : 92) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
CSRP2_MOUSE
|
||||||
| NC score | 0.020826 (rank : 93) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97314, Q9CZG5 | Gene names | Csrp2, Dlp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Double LIM protein 1) (DLP-1). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.020570 (rank : 94) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
GNAS1_HUMAN
|
||||||
| NC score | 0.020106 (rank : 95) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
PP2CG_HUMAN
|
||||||
| NC score | 0.018424 (rank : 96) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.018376 (rank : 97) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.018340 (rank : 98) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
PANK1_HUMAN
|
||||||
| NC score | 0.017956 (rank : 99) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
|
ARRD1_HUMAN
|
||||||
| NC score | 0.017625 (rank : 100) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
OAS3_HUMAN
|
||||||
| NC score | 0.017279 (rank : 101) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.016982 (rank : 102) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
SENP3_HUMAN
|
||||||
| NC score | 0.016494 (rank : 103) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H4L4, Q96PS4, Q9Y3W9 | Gene names | SENP3, SSP3, SUSP3 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.015923 (rank : 104) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
EPHA6_HUMAN
|
||||||
| NC score | 0.014606 (rank : 105) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UF33 | Gene names | EPHA6, EHK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephrin type-A receptor 6 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-2) (EPH homology kinase 2). | |||||
|
TLX3_HUMAN
|
||||||
| NC score | 0.013500 (rank : 106) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43711, Q96AD3 | Gene names | TLX3, HOX11L2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell leukemia homeobox protein 3 (Homeobox protein Hox-11L2). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.013402 (rank : 107) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
ANR38_HUMAN
|
||||||
| NC score | 0.013156 (rank : 108) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T7N3, Q6P9A0, Q86T71, Q86VE6, Q8NAX3 | Gene names | ANKRD38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.011628 (rank : 109) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
ZN516_HUMAN
|
||||||
| NC score | 0.011013 (rank : 110) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.010299 (rank : 111) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.009089 (rank : 112) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
WNT5A_MOUSE
|
||||||
| NC score | 0.008884 (rank : 113) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22725 | Gene names | Wnt5a, Wnt-5a | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-5a precursor. | |||||
|
WNT5A_HUMAN
|
||||||
| NC score | 0.008860 (rank : 114) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41221 | Gene names | WNT5A | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-5a precursor. | |||||
|
GP124_MOUSE
|
||||||
| NC score | 0.008525 (rank : 115) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.008080 (rank : 116) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
MK15_HUMAN
|
||||||
| NC score | 0.007948 (rank : 117) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
LAMB3_MOUSE
|
||||||
| NC score | 0.006336 (rank : 118) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
LAP2_HUMAN
|
||||||
| NC score | 0.005508 (rank : 119) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
ZN500_HUMAN
|
||||||
| NC score | 0.000550 (rank : 120) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60304, Q96CQ8, Q9BTG0 | Gene names | ZNF500, KIAA0557 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 500. | |||||