Please be patient as the page loads
|
LIPE_HUMAN
|
||||||
| SwissProt Accessions | Q9Y5X9, Q6UW82 | Gene names | LIPG | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL) (EL). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LIPE_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y5X9, Q6UW82 | Gene names | LIPG | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL) (EL). | |||||
|
LIPE_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998370 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVG5, Q8VDU2 | Gene names | Lipg | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL). | |||||
|
LIPL_HUMAN
|
||||||
| θ value | 4.02752e-125 (rank : 3) | NC score | 0.983650 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P06858, Q16282, Q16283, Q96FC4 | Gene names | LPL, LIPD | |||
|
Domain Architecture |

|
|||||
| Description | Lipoprotein lipase precursor (EC 3.1.1.34) (LPL). | |||||
|
LIPL_MOUSE
|
||||||
| θ value | 8.97236e-125 (rank : 4) | NC score | 0.984396 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11152, Q05956, Q9D3M9, Q9DCM8 | Gene names | Lpl | |||
|
Domain Architecture |

|
|||||
| Description | Lipoprotein lipase precursor (EC 3.1.1.34) (LPL). | |||||
|
LIPH_MOUSE
|
||||||
| θ value | 7.6132e-116 (rank : 5) | NC score | 0.980042 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27656 | Gene names | Lipc, Hpl | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic triacylglycerol lipase precursor (EC 3.1.1.3) (Hepatic lipase) (HL). | |||||
|
LIPH_HUMAN
|
||||||
| θ value | 4.61877e-113 (rank : 6) | NC score | 0.980241 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11150, O43571, P78529, Q99465 | Gene names | LIPC, HTGL | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic triacylglycerol lipase precursor (EC 3.1.1.3) (Hepatic lipase) (HL). | |||||
|
LIPR1_MOUSE
|
||||||
| θ value | 8.57503e-51 (rank : 7) | NC score | 0.891806 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5BKQ4, O70478, Q6NV82 | Gene names | Pnliprp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreatic lipase-related protein 1 precursor (EC 3.1.1.3). | |||||
|
LIPP_HUMAN
|
||||||
| θ value | 2.49495e-50 (rank : 8) | NC score | 0.891074 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16233 | Gene names | PNLIP | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic triacylglycerol lipase precursor (EC 3.1.1.3) (Pancreatic lipase) (PL). | |||||
|
LIPR1_HUMAN
|
||||||
| θ value | 1.04822e-48 (rank : 9) | NC score | 0.887972 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54315, Q68D83, Q68DR6, Q8TAU2, Q9BS82 | Gene names | PNLIPRP1, PLRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 1 precursor (EC 3.1.1.3). | |||||
|
LIPR2_HUMAN
|
||||||
| θ value | 1.36902e-48 (rank : 10) | NC score | 0.887532 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54317 | Gene names | PNLIPRP2, PLRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3). | |||||
|
LIPI_HUMAN
|
||||||
| θ value | 2.18568e-46 (rank : 11) | NC score | 0.925810 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6XZB0 | Gene names | LIPI, LPDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipase member I precursor (EC 3.1.1.-) (Membrane-associated phosphatidic acid-selective phospholipase A1-beta) (mPA-PLA1 beta) (LPD lipase). | |||||
|
LIPR2_MOUSE
|
||||||
| θ value | 2.95404e-43 (rank : 12) | NC score | 0.876564 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17892 | Gene names | Pnliprp2, Plrp2 | |||
|
Domain Architecture |
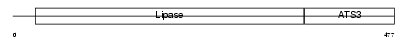
|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3) (Cytotoxic T-lymphocyte lipase). | |||||
|
PAPPA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.020699 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.046177 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.040547 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.048837 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
CMTA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.009470 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
RARG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.012941 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13631 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
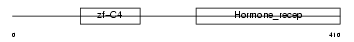
|
|||||
| Description | Retinoic acid receptor gamma-1 (RAR-gamma-1). | |||||
|
RARG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.012952 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18911, Q91VK5 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |
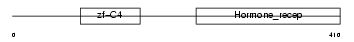
|
|||||
| Description | Retinoic acid receptor gamma-A (RAR-gamma-A). | |||||
|
RARG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.012913 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22932, Q15281 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor gamma-2 (RAR-gamma-2). | |||||
|
RARG2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.012910 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20787 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |
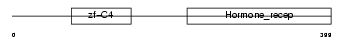
|
|||||
| Description | Retinoic acid receptor gamma-B (RAR-gamma-B). | |||||
|
RARA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.012375 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10276, P78456, Q13440, Q13441, Q9NQS0 | Gene names | RARA, NR1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
RARA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.012400 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11416, P22603 | Gene names | Rara, Nr1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
RARB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.012403 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10826, P12891, Q00989, Q15298, Q9UN48 | Gene names | RARB, HAP, NR1B2 | |||
|
Domain Architecture |
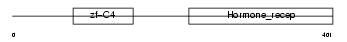
|
|||||
| Description | Retinoic acid receptor beta (RAR-beta) (RAR-epsilon) (HBV-activated protein). | |||||
|
RARB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.012402 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22605, P11417, P22604 | Gene names | Rarb, Nr1b2 | |||
|
Domain Architecture |
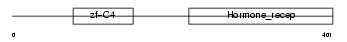
|
|||||
| Description | Retinoic acid receptor beta (RAR-beta). | |||||
|
LIPE_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y5X9, Q6UW82 | Gene names | LIPG | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL) (EL). | |||||
|
LIPE_MOUSE
|
||||||
| NC score | 0.998370 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVG5, Q8VDU2 | Gene names | Lipg | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL). | |||||
|
LIPL_MOUSE
|
||||||
| NC score | 0.984396 (rank : 3) | θ value | 8.97236e-125 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11152, Q05956, Q9D3M9, Q9DCM8 | Gene names | Lpl | |||
|
Domain Architecture |

|
|||||
| Description | Lipoprotein lipase precursor (EC 3.1.1.34) (LPL). | |||||
|
LIPL_HUMAN
|
||||||
| NC score | 0.983650 (rank : 4) | θ value | 4.02752e-125 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P06858, Q16282, Q16283, Q96FC4 | Gene names | LPL, LIPD | |||
|
Domain Architecture |

|
|||||
| Description | Lipoprotein lipase precursor (EC 3.1.1.34) (LPL). | |||||
|
LIPH_HUMAN
|
||||||
| NC score | 0.980241 (rank : 5) | θ value | 4.61877e-113 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11150, O43571, P78529, Q99465 | Gene names | LIPC, HTGL | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic triacylglycerol lipase precursor (EC 3.1.1.3) (Hepatic lipase) (HL). | |||||
|
LIPH_MOUSE
|
||||||
| NC score | 0.980042 (rank : 6) | θ value | 7.6132e-116 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27656 | Gene names | Lipc, Hpl | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic triacylglycerol lipase precursor (EC 3.1.1.3) (Hepatic lipase) (HL). | |||||
|
LIPI_HUMAN
|
||||||
| NC score | 0.925810 (rank : 7) | θ value | 2.18568e-46 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6XZB0 | Gene names | LIPI, LPDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipase member I precursor (EC 3.1.1.-) (Membrane-associated phosphatidic acid-selective phospholipase A1-beta) (mPA-PLA1 beta) (LPD lipase). | |||||
|
LIPR1_MOUSE
|
||||||
| NC score | 0.891806 (rank : 8) | θ value | 8.57503e-51 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5BKQ4, O70478, Q6NV82 | Gene names | Pnliprp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreatic lipase-related protein 1 precursor (EC 3.1.1.3). | |||||
|
LIPP_HUMAN
|
||||||
| NC score | 0.891074 (rank : 9) | θ value | 2.49495e-50 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16233 | Gene names | PNLIP | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic triacylglycerol lipase precursor (EC 3.1.1.3) (Pancreatic lipase) (PL). | |||||
|
LIPR1_HUMAN
|
||||||
| NC score | 0.887972 (rank : 10) | θ value | 1.04822e-48 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54315, Q68D83, Q68DR6, Q8TAU2, Q9BS82 | Gene names | PNLIPRP1, PLRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 1 precursor (EC 3.1.1.3). | |||||
|
LIPR2_HUMAN
|
||||||
| NC score | 0.887532 (rank : 11) | θ value | 1.36902e-48 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54317 | Gene names | PNLIPRP2, PLRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3). | |||||
|
LIPR2_MOUSE
|
||||||
| NC score | 0.876564 (rank : 12) | θ value | 2.95404e-43 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17892 | Gene names | Pnliprp2, Plrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3) (Cytotoxic T-lymphocyte lipase). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.048837 (rank : 13) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.046177 (rank : 14) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.040547 (rank : 15) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PAPPA_MOUSE
|
||||||
| NC score | 0.020699 (rank : 16) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
RARG1_MOUSE
|
||||||
| NC score | 0.012952 (rank : 17) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18911, Q91VK5 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-A (RAR-gamma-A). | |||||
|
RARG1_HUMAN
|
||||||
| NC score | 0.012941 (rank : 18) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13631 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-1 (RAR-gamma-1). | |||||
|
RARG2_HUMAN
|
||||||
| NC score | 0.012913 (rank : 19) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22932, Q15281 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor gamma-2 (RAR-gamma-2). | |||||
|
RARG2_MOUSE
|
||||||
| NC score | 0.012910 (rank : 20) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20787 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-B (RAR-gamma-B). | |||||
|
RARB_HUMAN
|
||||||
| NC score | 0.012403 (rank : 21) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10826, P12891, Q00989, Q15298, Q9UN48 | Gene names | RARB, HAP, NR1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor beta (RAR-beta) (RAR-epsilon) (HBV-activated protein). | |||||
|
RARB_MOUSE
|
||||||
| NC score | 0.012402 (rank : 22) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22605, P11417, P22604 | Gene names | Rarb, Nr1b2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor beta (RAR-beta). | |||||
|
RARA_MOUSE
|
||||||
| NC score | 0.012400 (rank : 23) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11416, P22603 | Gene names | Rara, Nr1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
RARA_HUMAN
|
||||||
| NC score | 0.012375 (rank : 24) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10276, P78456, Q13440, Q13441, Q9NQS0 | Gene names | RARA, NR1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
CMTA2_MOUSE
|
||||||
| NC score | 0.009470 (rank : 25) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||