Please be patient as the page loads
|
HIF1A_HUMAN
|
||||||
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HIF1A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993764 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 3.1544e-162 (rank : 3) | NC score | 0.976450 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 2.04462e-161 (rank : 4) | NC score | 0.975288 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 9.41505e-74 (rank : 5) | NC score | 0.933296 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 3.57772e-73 (rank : 6) | NC score | 0.932070 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| θ value | 5.1662e-72 (rank : 7) | NC score | 0.930358 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 5.1662e-72 (rank : 8) | NC score | 0.929965 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 9) | NC score | 0.905775 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 8.55514e-59 (rank : 10) | NC score | 0.905553 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | 6.56568e-51 (rank : 11) | NC score | 0.906437 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 1.04822e-48 (rank : 12) | NC score | 0.906407 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 1.37539e-32 (rank : 13) | NC score | 0.824301 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 1.37539e-32 (rank : 14) | NC score | 0.822124 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 15) | NC score | 0.803915 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 3.17079e-29 (rank : 16) | NC score | 0.800620 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 17) | NC score | 0.783999 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 7.06379e-29 (rank : 18) | NC score | 0.789520 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 2.27234e-27 (rank : 19) | NC score | 0.823816 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 20) | NC score | 0.775330 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 5.06226e-27 (rank : 21) | NC score | 0.762996 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 8.63488e-27 (rank : 22) | NC score | 0.826852 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 23) | NC score | 0.780297 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 24) | NC score | 0.779499 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 25) | NC score | 0.675250 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 26) | NC score | 0.656738 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 27) | NC score | 0.566777 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 28) | NC score | 0.622421 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 29) | NC score | 0.624412 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 30) | NC score | 0.566636 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 31) | NC score | 0.565466 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 32) | NC score | 0.529163 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 33) | NC score | 0.338288 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 34) | NC score | 0.316526 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 35) | NC score | 0.313510 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 36) | NC score | 0.303804 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 37) | NC score | 0.308402 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
SN1L2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 38) | NC score | 0.003544 (rank : 120) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 39) | NC score | 0.285404 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 40) | NC score | 0.040290 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
HCN1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 41) | NC score | 0.014898 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.020047 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.019095 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
RIF1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.022135 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.029222 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.056982 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.013438 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
GPR64_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.009452 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CJ12, Q8BL10, Q8CJ08, Q8CJ09, Q8CJ10 | Gene names | Gpr64, Me6 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (Me6 receptor). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.022139 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.018226 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
STON2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.018980 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.031600 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.015031 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.009977 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.009791 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
S12A7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.007585 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.009324 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
APBA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.008984 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
GP156_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.016761 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
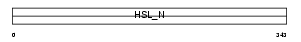
LIPS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.016478 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54310, P97866, Q3TE34, Q6GU16 | Gene names | Lipe | |||
|
Domain Architecture |
|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.012204 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
CJ047_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.017511 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
DDHD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.019565 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.014099 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.014703 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.018285 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
HES1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.033216 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.033449 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.014045 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.009955 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.039779 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.013384 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CENPT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.015770 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.025507 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.000424 (rank : 128) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | -0.000653 (rank : 133) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HYES_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.012019 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34914, Q8CGV0 | Gene names | Ephx2, Eph2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
|
LRRC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.000813 (rank : 126) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBW1, Q6ZMI8, Q96A85 | Gene names | LRRC4, BAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 4 precursor (Brain tumor- associated protein LRRC4) (NAG14). | |||||
|
PALM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.012160 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
PHF13_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.012909 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YI8, Q8N551, Q9UJP2 | Gene names | PHF13 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 13. | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.013438 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.018454 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.027499 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.007456 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
ATS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.001135 (rank : 125) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97857, O54768 | Gene names | Adamts1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1). | |||||
|
DOK4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.010237 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TEW6, O75209, Q9BTP2, Q9NVV3 | Gene names | DOK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.010259 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KE3, Q3U0X3 | Gene names | Dok4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
IF16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.009115 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.019184 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PACS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.011837 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
PASK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.002822 (rank : 122) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.023960 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SN1L2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | -0.000594 (rank : 132) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
TRI47_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.005670 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0E3, Q6P249, Q811J7, Q8BVZ8, Q8R1K0, Q8R3Y1 | Gene names | Trim47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47. | |||||
|
ZN410_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | -0.003887 (rank : 134) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86VK4, O00153, Q9BQ19 | Gene names | ZNF410, APA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 410 (Zinc finger protein APA-1) (Another partner for ARF 1). | |||||
|
AP2A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.004387 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
BCL7B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.008766 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921K9, O89022, Q3TV31, Q3U2W0 | Gene names | Bcl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.018302 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CF152_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.003682 (rank : 119) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
CMTA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.008765 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
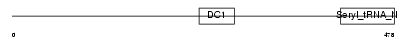
DTNA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.004394 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |
|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.004727 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
LRRC7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.000233 (rank : 130) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.001481 (rank : 124) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.008220 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
ROBO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.000349 (rank : 129) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
SNPC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.010620 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13487, Q13486 | Gene names | SNAPC2, SNAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2) (Proximal sequence element-binding transcription factor subunit delta) (PSE- binding factor subunit delta) (PTF subunit delta). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.016777 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
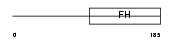
FOXF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.000805 (rank : 127) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12947, Q9UQ85 | Gene names | FOXF2, FKHL6, FREAC2 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein F2 (Forkhead-related protein FKHL6) (Forkhead- related transcription factor 2) (FREAC-2) (Forkhead-related activator 2). | |||||
|
G3BP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.003325 (rank : 121) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
PDIP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.008763 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG81, Q6PE83, Q80X99, Q8CI28 | Gene names | Poldip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polymerase delta-interacting protein 3. | |||||
|
PLCB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.002718 (rank : 123) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PP1RA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.006405 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
PP4R1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.005868 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
ROBO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.000101 (rank : 131) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.031561 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.005083 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
ZN358_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | -0.004702 (rank : 135) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NW07, Q9BTM7 | Gene names | ZNF358 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 358. | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.064582 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.083500 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.087321 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.093977 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.091866 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.073595 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.075304 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.065863 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.065831 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.075848 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.075204 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.070036 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.069185 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.068423 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.068392 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.065699 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.061024 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.993764 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.976450 (rank : 3) | θ value | 3.1544e-162 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.975288 (rank : 4) | θ value | 2.04462e-161 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.933296 (rank : 5) | θ value | 9.41505e-74 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.932070 (rank : 6) | θ value | 3.57772e-73 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.930358 (rank : 7) | θ value | 5.1662e-72 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.929965 (rank : 8) | θ value | 5.1662e-72 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.906437 (rank : 9) | θ value | 6.56568e-51 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.906407 (rank : 10) | θ value | 1.04822e-48 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.905775 (rank : 11) | θ value | 8.55514e-59 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.905553 (rank : 12) | θ value | 8.55514e-59 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.826852 (rank : 13) | θ value | 8.63488e-27 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.824301 (rank : 14) | θ value | 1.37539e-32 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.823816 (rank : 15) | θ value | 2.27234e-27 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.822124 (rank : 16) | θ value | 1.37539e-32 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.803915 (rank : 17) | θ value | 8.3442e-30 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.800620 (rank : 18) | θ value | 3.17079e-29 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.789520 (rank : 19) | θ value | 7.06379e-29 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.783999 (rank : 20) | θ value | 7.06379e-29 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.780297 (rank : 21) | θ value | 4.28545e-26 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.779499 (rank : 22) | θ value | 5.59698e-26 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.775330 (rank : 23) | θ value | 3.87602e-27 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.762996 (rank : 24) | θ value | 5.06226e-27 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.675250 (rank : 25) | θ value | 2.35696e-16 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.656738 (rank : 26) | θ value | 3.76295e-14 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.624412 (rank : 27) | θ value | 5.43371e-13 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.622421 (rank : 28) | θ value | 3.18553e-13 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.566777 (rank : 29) | θ value | 1.42992e-13 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.566636 (rank : 30) | θ value | 9.26847e-13 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.565466 (rank : 31) | θ value | 1.47974e-10 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.529163 (rank : 32) | θ value | 6.21693e-09 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.338288 (rank : 33) | θ value | 6.43352e-06 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.316526 (rank : 34) | θ value | 1.43324e-05 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.313510 (rank : 35) | θ value | 1.43324e-05 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.308402 (rank : 36) | θ value | 0.00228821 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.303804 (rank : 37) | θ value | 0.000461057 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.285404 (rank : 38) | θ value | 0.0113563 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.093977 (rank : 39) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.091866 (rank : 40) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.087321 (rank : 41) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.083500 (rank : 42) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.075848 (rank : 43) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.075304 (rank : 44) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.075204 (rank : 45) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.073595 (rank : 46) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.070036 (rank : 47) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.069185 (rank : 48) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.068423 (rank : 49) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.068392 (rank : 50) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.065863 (rank : 51) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.065831 (rank : 52) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.065699 (rank : 53) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.064582 (rank : 54) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.061024 (rank : 55) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.056982 (rank : 56) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.040290 (rank : 57) | θ value | 0.0252991 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.039779 (rank : 58) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.033449 (rank : 59) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.033216 (rank : 60) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.031600 (rank : 61) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.031561 (rank : 62) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.029222 (rank : 63) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.027499 (rank : 64) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.025507 (rank : 65) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.023960 (rank : 66) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.022139 (rank : 67) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
RIF1_HUMAN
|
||||||
| NC score | 0.022135 (rank : 68) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.020047 (rank : 69) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
DDHD1_HUMAN
|
||||||
| NC score | 0.019565 (rank : 70) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.019184 (rank : 71) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.019095 (rank : 72) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
STON2_HUMAN
|
||||||
| NC score | 0.018980 (rank : 73) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.018454 (rank : 74) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.018302 (rank : 75) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.018285 (rank : 76) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.018226 (rank : 77) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
CJ047_MOUSE
|
||||||
| NC score | 0.017511 (rank : 78) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.016777 (rank : 79) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
GP156_MOUSE
|
||||||
| NC score | 0.016761 (rank : 80) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
LIPS_MOUSE
|
||||||
| NC score | 0.016478 (rank : 81) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54310, P97866, Q3TE34, Q6GU16 | Gene names | Lipe | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
CENPT_MOUSE
|
||||||
| NC score | 0.015770 (rank : 82) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.015031 (rank : 83) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.014898 (rank : 84) | θ value | 0.0736092 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.014703 (rank : 85) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.014099 (rank : 86) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.014045 (rank : 87) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.013438 (rank : 88) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.013438 (rank : 89) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.013384 (rank : 90) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
PHF13_HUMAN
|
||||||
| NC score | 0.012909 (rank : 91) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YI8, Q8N551, Q9UJP2 | Gene names | PHF13 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 13. | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.012204 (rank : 92) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.012160 (rank : 93) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
HYES_MOUSE
|
||||||
| NC score | 0.012019 (rank : 94) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34914, Q8CGV0 | Gene names | Ephx2, Eph2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
|
PACS1_MOUSE
|
||||||
| NC score | 0.011837 (rank : 95) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
SNPC2_HUMAN
|
||||||
| NC score | 0.010620 (rank : 96) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13487, Q13486 | Gene names | SNAPC2, SNAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2) (Proximal sequence element-binding transcription factor subunit delta) (PSE- binding factor subunit delta) (PTF subunit delta). | |||||
|
DOK4_MOUSE
|
||||||
| NC score | 0.010259 (rank : 97) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KE3, Q3U0X3 | Gene names | Dok4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK4_HUMAN
|
||||||
| NC score | 0.010237 (rank : 98) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TEW6, O75209, Q9BTP2, Q9NVV3 | Gene names | DOK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.009977 (rank : 99) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.009955 (rank : 100) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.009791 (rank : 101) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
GPR64_MOUSE
|
||||||
| NC score | 0.009452 (rank : 102) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CJ12, Q8BL10, Q8CJ08, Q8CJ09, Q8CJ10 | Gene names | Gpr64, Me6 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (Me6 receptor). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.009324 (rank : 103) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.009115 (rank : 104) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.008984 (rank : 105) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
BCL7B_MOUSE
|
||||||
| NC score | 0.008766 (rank : 106) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921K9, O89022, Q3TV31, Q3U2W0 | Gene names | Bcl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
CMTA1_HUMAN
|
||||||
| NC score | 0.008765 (rank : 107) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
PDIP3_MOUSE
|
||||||
| NC score | 0.008763 (rank : 108) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG81, Q6PE83, Q80X99, Q8CI28 | Gene names | Poldip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polymerase delta-interacting protein 3. | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.008220 (rank : 109) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
S12A7_MOUSE
|
||||||
| NC score | 0.007585 (rank : 110) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.007456 (rank : 111) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
PP1RA_MOUSE
|
||||||
| NC score | 0.006405 (rank : 112) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
PP4R1_MOUSE
|
||||||
| NC score | 0.005868 (rank : 113) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
TRI47_MOUSE
|
||||||
| NC score | 0.005670 (rank : 114) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0E3, Q6P249, Q811J7, Q8BVZ8, Q8R1K0, Q8R3Y1 | Gene names | Trim47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47. | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.005083 (rank : 115) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.004727 (rank : 116) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
DTNA_MOUSE
|
||||||
| NC score | 0.004394 (rank : 117) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
AP2A1_HUMAN
|
||||||
| NC score | 0.004387 (rank : 118) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.003682 (rank : 119) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
SN1L2_HUMAN
|
||||||
| NC score | 0.003544 (rank : 120) | θ value | 0.00869519 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
G3BP2_MOUSE
|
||||||
| NC score | 0.003325 (rank : 121) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
PASK_MOUSE
|
||||||
| NC score | 0.002822 (rank : 122) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||
|
PLCB1_MOUSE
|
||||||
| NC score | 0.002718 (rank : 123) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.001481 (rank : 124) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
ATS1_MOUSE
|
||||||
| NC score | 0.001135 (rank : 125) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97857, O54768 | Gene names | Adamts1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1). | |||||
|
LRRC4_HUMAN
|
||||||
| NC score | 0.000813 (rank : 126) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBW1, Q6ZMI8, Q96A85 | Gene names | LRRC4, BAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 4 precursor (Brain tumor- associated protein LRRC4) (NAG14). | |||||
|
FOXF2_HUMAN
|
||||||
| NC score | 0.000805 (rank : 127) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12947, Q9UQ85 | Gene names | FOXF2, FKHL6, FREAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein F2 (Forkhead-related protein FKHL6) (Forkhead- related transcription factor 2) (FREAC-2) (Forkhead-related activator 2). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.000424 (rank : 128) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
ROBO1_MOUSE
|
||||||
| NC score | 0.000349 (rank : 129) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
LRRC7_HUMAN
|
||||||
| NC score | 0.000233 (rank : 130) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
ROBO1_HUMAN
|
||||||
| NC score | 0.000101 (rank : 131) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
SN1L2_MOUSE
|
||||||
| NC score | -0.000594 (rank : 132) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | -0.000653 (rank : 133) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
ZN410_HUMAN
|
||||||
| NC score | -0.003887 (rank : 134) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86VK4, O00153, Q9BQ19 | Gene names | ZNF410, APA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 410 (Zinc finger protein APA-1) (Another partner for ARF 1). | |||||
|
ZN358_HUMAN
|
||||||
| NC score | -0.004702 (rank : 135) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NW07, Q9BTM7 | Gene names | ZNF358 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 358. | |||||