Please be patient as the page loads
|
SIM2_MOUSE
|
||||||
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SIM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987961 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987324 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.995700 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 2.0926e-81 (rank : 5) | NC score | 0.943133 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 4.66183e-81 (rank : 6) | NC score | 0.944354 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 1.5519e-76 (rank : 7) | NC score | 0.952717 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | 5.51964e-74 (rank : 8) | NC score | 0.951467 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 7.20884e-74 (rank : 9) | NC score | 0.935205 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 3.57772e-73 (rank : 10) | NC score | 0.932070 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 7.98882e-65 (rank : 11) | NC score | 0.924058 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 2.32439e-64 (rank : 12) | NC score | 0.923554 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 1.85889e-29 (rank : 13) | NC score | 0.837946 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 2.68423e-28 (rank : 14) | NC score | 0.791611 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 4.57862e-28 (rank : 15) | NC score | 0.838702 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 16) | NC score | 0.793017 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 17) | NC score | 0.703375 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 18) | NC score | 0.747697 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 19) | NC score | 0.746194 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 20) | NC score | 0.759126 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 4.9032e-22 (rank : 21) | NC score | 0.756072 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 22) | NC score | 0.723508 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 23) | NC score | 0.735500 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 1.33526e-19 (rank : 24) | NC score | 0.686735 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 25) | NC score | 0.734706 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 26) | NC score | 0.636165 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 27) | NC score | 0.726641 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 28) | NC score | 0.632476 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 29) | NC score | 0.557589 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 30) | NC score | 0.555783 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 31) | NC score | 0.555552 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 32) | NC score | 0.519152 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 33) | NC score | 0.210808 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
SCND1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 34) | NC score | 0.019841 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57086 | Gene names | SCAND1, SDP1 | |||
|
Domain Architecture |
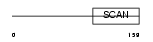
|
|||||
| Description | SCAN domain-containing protein 1. | |||||
|
TBCD1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 35) | NC score | 0.026576 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
US6NL_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 36) | NC score | 0.035853 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 37) | NC score | 0.230988 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.022908 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.215369 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.018336 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.019895 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.239970 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
EXOC3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.029849 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6KAR6 | Gene names | Exoc3, Sec6l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
|
FOXO1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.010705 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12778, O43523, Q5VYC7, Q6NSK6 | Gene names | FOXO1A, FKHR | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O1A (Forkhead in rhabdomyosarcoma). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.013208 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.028980 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
HNF3B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.006812 (rank : 78) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35583, Q60602 | Gene names | Foxa2, Hnf3b, Tcf-3b, Tcf3b | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-beta (HNF-3B) (Forkhead box protein A2). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.113296 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.111543 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
JHD2B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.011758 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.008565 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.012492 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.006911 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
DC1L2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.011497 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.102612 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.106259 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
LTB1L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.003521 (rank : 86) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.003539 (rank : 85) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
DC12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.014806 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96FZ2, Q96G34, Q9NRP3 | Gene names | C3orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0361 protein DC12. | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.006296 (rank : 79) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
CX045_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.017696 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H5U8, Q5JXY9 | Gene names | CXorf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CXorf45. | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.006212 (rank : 80) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
NRIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.010999 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48552, Q8IWE8 | Gene names | NRIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
GLIS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | -0.003242 (rank : 89) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NBF1, Q8N9V9 | Gene names | GLIS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | -0.003696 (rank : 90) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
MEF2B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.007722 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
EXOC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.016811 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60645, Q8TEN6, Q8WUW0, Q96DI4 | Gene names | EXOC3, SEC6, SEC6L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
|
HXB9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | -0.000317 (rank : 88) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17482, Q9H1I1 | Gene names | HOXB9, HOX2E | |||
|
Domain Architecture |
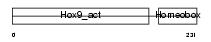
|
|||||
| Description | Homeobox protein Hox-B9 (Hox-2E) (Hox-2.5). | |||||
|
NFAC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.005620 (rank : 82) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.003701 (rank : 84) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
TAL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.010249 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
HNF3B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.004527 (rank : 83) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y261, Q8WUW4, Q96DF7 | Gene names | FOXA2, HNF3B, TCF3B | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-beta (HNF-3B) (Forkhead box protein A2). | |||||
|
K0528_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.005960 (rank : 81) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YS7, O60280, Q7Z619, Q86SU3 | Gene names | KIAA0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.007450 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
TLE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.000981 (rank : 87) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62440 | Gene names | Tle1, Grg1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (Groucho-related protein 1) (Grg- 1). | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.069140 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.068644 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.070131 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.064261 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.064308 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
PER1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.215064 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.211428 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.068799 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.068024 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.064606 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.064469 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.063887 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.063858 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.061674 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.057653 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.995700 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.987961 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.987324 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.952717 (rank : 5) | θ value | 1.5519e-76 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.951467 (rank : 6) | θ value | 5.51964e-74 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.944354 (rank : 7) | θ value | 4.66183e-81 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.943133 (rank : 8) | θ value | 2.0926e-81 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.935205 (rank : 9) | θ value | 7.20884e-74 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.932070 (rank : 10) | θ value | 3.57772e-73 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.924058 (rank : 11) | θ value | 7.98882e-65 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.923554 (rank : 12) | θ value | 2.32439e-64 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.838702 (rank : 13) | θ value | 4.57862e-28 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.837946 (rank : 14) | θ value | 1.85889e-29 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.793017 (rank : 15) | θ value | 2.27234e-27 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.791611 (rank : 16) | θ value | 2.68423e-28 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.759126 (rank : 17) | θ value | 2.87452e-22 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.756072 (rank : 18) | θ value | 4.9032e-22 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.747697 (rank : 19) | θ value | 1.16704e-23 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.746194 (rank : 20) | θ value | 1.16704e-23 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.735500 (rank : 21) | θ value | 1.2077e-20 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.734706 (rank : 22) | θ value | 1.92812e-18 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.726641 (rank : 23) | θ value | 3.63628e-17 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.723508 (rank : 24) | θ value | 1.42661e-21 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.703375 (rank : 25) | θ value | 8.93572e-24 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.686735 (rank : 26) | θ value | 1.33526e-19 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.636165 (rank : 27) | θ value | 5.60996e-18 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.632476 (rank : 28) | θ value | 8.10077e-17 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.557589 (rank : 29) | θ value | 5.08577e-11 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.555783 (rank : 30) | θ value | 3.29651e-10 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.555552 (rank : 31) | θ value | 4.30538e-10 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.519152 (rank : 32) | θ value | 1.17247e-07 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.239970 (rank : 33) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.230988 (rank : 34) | θ value | 0.0736092 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.215369 (rank : 35) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.215064 (rank : 36) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.211428 (rank : 37) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.210808 (rank : 38) | θ value | 0.0193708 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.113296 (rank : 39) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.111543 (rank : 40) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.106259 (rank : 41) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.102612 (rank : 42) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.070131 (rank : 43) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.069140 (rank : 44) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.068799 (rank : 45) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.068644 (rank : 46) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.068024 (rank : 47) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.064606 (rank : 48) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.064469 (rank : 49) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.064308 (rank : 50) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.064261 (rank : 51) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.063887 (rank : 52) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.063858 (rank : 53) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.061674 (rank : 54) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.057653 (rank : 55) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
US6NL_HUMAN
|
||||||
| NC score | 0.035853 (rank : 56) | θ value | 0.0431538 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
EXOC3_MOUSE
|
||||||
| NC score | 0.029849 (rank : 57) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6KAR6 | Gene names | Exoc3, Sec6l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.028980 (rank : 58) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
TBCD1_MOUSE
|
||||||
| NC score | 0.026576 (rank : 59) | θ value | 0.0330416 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.022908 (rank : 60) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.019895 (rank : 61) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
SCND1_HUMAN
|
||||||
| NC score | 0.019841 (rank : 62) | θ value | 0.0193708 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57086 | Gene names | SCAND1, SDP1 | |||
|
Domain Architecture |

|
|||||
| Description | SCAN domain-containing protein 1. | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.018336 (rank : 63) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
CX045_HUMAN
|
||||||
| NC score | 0.017696 (rank : 64) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H5U8, Q5JXY9 | Gene names | CXorf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CXorf45. | |||||
|
EXOC3_HUMAN
|
||||||
| NC score | 0.016811 (rank : 65) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60645, Q8TEN6, Q8WUW0, Q96DI4 | Gene names | EXOC3, SEC6, SEC6L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
|
DC12_HUMAN
|
||||||
| NC score | 0.014806 (rank : 66) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96FZ2, Q96G34, Q9NRP3 | Gene names | C3orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0361 protein DC12. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.013208 (rank : 67) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.012492 (rank : 68) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
JHD2B_MOUSE
|
||||||
| NC score | 0.011758 (rank : 69) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
DC1L2_MOUSE
|
||||||
| NC score | 0.011497 (rank : 70) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
NRIP1_HUMAN
|
||||||
| NC score | 0.010999 (rank : 71) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48552, Q8IWE8 | Gene names | NRIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
FOXO1_HUMAN
|
||||||
| NC score | 0.010705 (rank : 72) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12778, O43523, Q5VYC7, Q6NSK6 | Gene names | FOXO1A, FKHR | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O1A (Forkhead in rhabdomyosarcoma). | |||||
|
TAL1_HUMAN
|
||||||
| NC score | 0.010249 (rank : 73) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.008565 (rank : 74) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
MEF2B_HUMAN
|
||||||
| NC score | 0.007722 (rank : 75) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.007450 (rank : 76) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.006911 (rank : 77) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
HNF3B_MOUSE
|
||||||
| NC score | 0.006812 (rank : 78) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35583, Q60602 | Gene names | Foxa2, Hnf3b, Tcf-3b, Tcf3b | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-beta (HNF-3B) (Forkhead box protein A2). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.006296 (rank : 79) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.006212 (rank : 80) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
K0528_HUMAN
|
||||||
| NC score | 0.005960 (rank : 81) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YS7, O60280, Q7Z619, Q86SU3 | Gene names | KIAA0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.005620 (rank : 82) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
HNF3B_HUMAN
|
||||||
| NC score | 0.004527 (rank : 83) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y261, Q8WUW4, Q96DF7 | Gene names | FOXA2, HNF3B, TCF3B | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-beta (HNF-3B) (Forkhead box protein A2). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.003701 (rank : 84) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
LTB1S_HUMAN
|
||||||
| NC score | 0.003539 (rank : 85) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1L_HUMAN
|
||||||
| NC score | 0.003521 (rank : 86) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
TLE1_MOUSE
|
||||||
| NC score | 0.000981 (rank : 87) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62440 | Gene names | Tle1, Grg1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (Groucho-related protein 1) (Grg- 1). | |||||
|
HXB9_HUMAN
|
||||||
| NC score | -0.000317 (rank : 88) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17482, Q9H1I1 | Gene names | HOXB9, HOX2E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B9 (Hox-2E) (Hox-2.5). | |||||
|
GLIS1_HUMAN
|
||||||
| NC score | -0.003242 (rank : 89) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NBF1, Q8N9V9 | Gene names | GLIS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | -0.003696 (rank : 90) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||