Please be patient as the page loads
|
MLX_MOUSE
|
||||||
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MLX_MOUSE
|
||||||
| θ value | 9.19922e-154 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| θ value | 2.42647e-146 (rank : 2) | NC score | 0.995677 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 3) | NC score | 0.558132 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 4) | NC score | 0.526280 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 5) | NC score | 0.360723 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 6) | NC score | 0.471341 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
USF1_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 7) | NC score | 0.432230 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 8) | NC score | 0.432604 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 9) | NC score | 0.358160 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
MAX_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 10) | NC score | 0.461639 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 11) | NC score | 0.351072 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
USF2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 12) | NC score | 0.421414 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 13) | NC score | 0.425036 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 14) | NC score | 0.224782 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 15) | NC score | 0.284117 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 16) | NC score | 0.289732 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 17) | NC score | 0.223472 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 18) | NC score | 0.321065 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 19) | NC score | 0.320280 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.196144 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 21) | NC score | 0.286660 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYC_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 22) | NC score | 0.289489 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 23) | NC score | 0.325189 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MYC_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 24) | NC score | 0.289930 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 25) | NC score | 0.192989 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.036859 (rank : 128) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
HEN2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.135634 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |
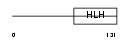
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.204771 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
HEN2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.133771 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |
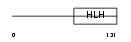
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 30) | NC score | 0.046095 (rank : 125) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.235145 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.255636 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.270075 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
ERBB3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 34) | NC score | 0.015093 (rank : 155) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
MITF_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.280055 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
HEN1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 36) | NC score | 0.124480 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |
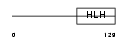
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.232636 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.241411 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.205543 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.204986 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
FIGLA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.147954 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
MITF_MOUSE
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.274097 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.029613 (rank : 142) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.159794 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.159039 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
PHAR2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.057834 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.185586 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.027522 (rank : 145) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.237695 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.045910 (rank : 126) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
TAL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.113339 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
TAL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.112166 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.176044 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
FIGLA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.146946 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
ERBB3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.012421 (rank : 158) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.030947 (rank : 137) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.039850 (rank : 127) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.029658 (rank : 141) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.025962 (rank : 148) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
ITF2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.048880 (rank : 123) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
LAMB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.011064 (rank : 161) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.093251 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.093059 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
PLXD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.018641 (rank : 154) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.028139 (rank : 143) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
ID1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.089102 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41134, O00651, O00652, Q16371, Q16377, Q5TE66, Q5TE67, Q969Z7, Q9H0Z5, Q9H109 | Gene names | ID1, ID | |||
|
Domain Architecture |
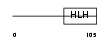
|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
ID1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.092156 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P20067, Q61101, Q9D897 | Gene names | Id1, Id, Id-1, Idb1 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
TULP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.030106 (rank : 139) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
CLK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.004127 (rank : 162) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49759 | Gene names | CLK1, CLK | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase CLK1 (EC 2.7.12.1) (CDC-like kinase 1). | |||||
|
HEN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.095645 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.033988 (rank : 133) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
NGN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.047019 (rank : 124) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.030389 (rank : 138) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.029704 (rank : 140) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
CCD11_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.028002 (rank : 144) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D439 | Gene names | Ccdc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
HOME2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.035324 (rank : 131) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NSB8, O95269, O95349, Q9NSB6, Q9NSB7, Q9UNT7 | Gene names | HOMER2 | |||
|
Domain Architecture |
|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.026236 (rank : 147) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
TRAK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.035484 (rank : 130) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |
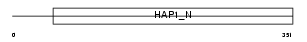
|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.160457 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.161243 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.168694 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.166800 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
CDC37_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.033953 (rank : 134) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |
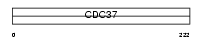
|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.011732 (rank : 160) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.033409 (rank : 135) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.034192 (rank : 132) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
MYF5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.053839 (rank : 113) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |
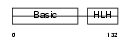
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.053610 (rank : 115) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |
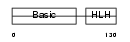
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
PCDGM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.001649 (rank : 163) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5F6, Q9Y5C2 | Gene names | PCDHGC5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C5 precursor (PCDH-gamma-C5). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.013041 (rank : 157) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
XLR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.035703 (rank : 129) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05531 | Gene names | Xlr | |||
|
Domain Architecture |
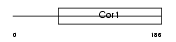
|
|||||
| Description | X-linked lymphocyte-regulated protein PM1. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.024434 (rank : 151) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.011986 (rank : 159) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CV019_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.033079 (rank : 136) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13769, O60839, Q9UPZ5 | Gene names | C22orf19, KIAA0983 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C22orf19 (NF2/meningioma region protein pK1.3) (Placental protein 39.2). | |||||
|
MFAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.025348 (rank : 150) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CQU1, Q3TU29, Q8CCL1, Q9CSJ5 | Gene names | Mfap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microfibrillar-associated protein 1. | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.022960 (rank : 152) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
CCD27_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.026874 (rank : 146) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.025520 (rank : 149) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.022892 (rank : 153) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SAS10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.013272 (rank : 156) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQZ2, Q6FI82 | Gene names | CRLZ1, SAS10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1). | |||||
|
AHR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.066840 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.065402 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
ASCL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.053840 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
ASCL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.050592 (rank : 121) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
ASCL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.050823 (rank : 120) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
BHLH8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.080252 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.080931 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.067571 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.066095 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
HAND1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.068336 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |
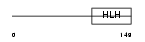
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
HAND1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.065468 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
HAND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.074297 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |
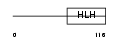
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.074297 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
HES1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.129233 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.129535 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.084665 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.089537 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.081597 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
HES4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.135424 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.123772 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HES6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.089538 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
HES6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.087281 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
HES7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.077041 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.076462 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.065831 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.065290 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
LYL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.061719 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
LYL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.059426 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
MAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.063529 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.080152 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.074460 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.089160 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.053711 (rank : 114) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.105122 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.103313 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.087053 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.091477 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MYCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.116107 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.166920 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.056464 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.056864 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.060957 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.054624 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.053426 (rank : 116) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.055194 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.058502 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.059552 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.052586 (rank : 118) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
PER2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.050227 (rank : 122) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PTF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.063103 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
PTF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.061716 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
SCX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.052729 (rank : 117) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |
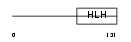
|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
SIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.063645 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.063902 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.064860 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.064308 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.216353 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
TAL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.050996 (rank : 119) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
TAL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.058048 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
TWST1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.064722 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.064645 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
TWST2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.063723 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.063821 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 9.19922e-154 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.995677 (rank : 2) | θ value | 2.42647e-146 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.558132 (rank : 3) | θ value | 5.07402e-19 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.526280 (rank : 4) | θ value | 4.29542e-18 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.471341 (rank : 5) | θ value | 3.77169e-06 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.461639 (rank : 6) | θ value | 1.43324e-05 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.432604 (rank : 7) | θ value | 6.43352e-06 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.432230 (rank : 8) | θ value | 6.43352e-06 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.425036 (rank : 9) | θ value | 0.000121331 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.421414 (rank : 10) | θ value | 0.000121331 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.360723 (rank : 11) | θ value | 2.21117e-06 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.358160 (rank : 12) | θ value | 8.40245e-06 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.351072 (rank : 13) | θ value | 9.29e-05 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.325189 (rank : 14) | θ value | 0.00390308 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.321065 (rank : 15) | θ value | 0.00102713 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.320280 (rank : 16) | θ value | 0.00134147 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.289930 (rank : 17) | θ value | 0.00509761 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.289732 (rank : 18) | θ value | 0.000786445 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.289489 (rank : 19) | θ value | 0.00390308 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.286660 (rank : 20) | θ value | 0.00390308 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.284117 (rank : 21) | θ value | 0.000461057 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.280055 (rank : 22) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.274097 (rank : 23) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.270075 (rank : 24) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.255636 (rank : 25) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.241411 (rank : 26) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.237695 (rank : 27) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.235145 (rank : 28) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.232636 (rank : 29) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.224782 (rank : 30) | θ value | 0.000461057 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.223472 (rank : 31) | θ value | 0.00102713 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.216353 (rank : 32) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.205543 (rank : 33) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.204986 (rank : 34) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.204771 (rank : 35) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.196144 (rank : 36) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.192989 (rank : 37) | θ value | 0.00665767 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.185586 (rank : 38) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.176044 (rank : 39) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.168694 (rank : 40) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.166920 (rank : 41) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.166800 (rank : 42) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.161243 (rank : 43) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.160457 (rank : 44) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.159794 (rank : 45) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.159039 (rank : 46) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
FIGLA_HUMAN
|
||||||
| NC score | 0.147954 (rank : 47) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
FIGLA_MOUSE
|
||||||
| NC score | 0.146946 (rank : 48) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
HEN2_HUMAN
|
||||||
| NC score | 0.135634 (rank : 49) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.135424 (rank : 50) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
HEN2_MOUSE
|
||||||
| NC score | 0.133771 (rank : 51) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.129535 (rank : 52) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.129233 (rank : 53) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HEN1_MOUSE
|
||||||
| NC score | 0.124480 (rank : 54) | θ value | 0.21417 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.123772 (rank : 55) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
MYCB_MOUSE
|
||||||
| NC score | 0.116107 (rank : 56) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
TAL1_HUMAN
|
||||||
| NC score | 0.113339 (rank : 57) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
TAL1_MOUSE
|
||||||
| NC score | 0.112166 (rank : 58) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.105122 (rank : 59) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.103313 (rank : 60) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
HEN1_HUMAN
|
||||||
| NC score | 0.095645 (rank : 61) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.093251 (rank : 62) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.093059 (rank : 63) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
ID1_MOUSE
|
||||||
| NC score | 0.092156 (rank : 64) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P20067, Q61101, Q9D897 | Gene names | Id1, Id, Id-1, Idb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.091477 (rank : 65) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
HES6_HUMAN
|
||||||
| NC score | 0.089538 (rank : 66) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
HES2_MOUSE
|
||||||
| NC score | 0.089537 (rank : 67) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.089160 (rank : 68) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
ID1_HUMAN
|
||||||
| NC score | 0.089102 (rank : 69) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41134, O00651, O00652, Q16371, Q16377, Q5TE66, Q5TE67, Q969Z7, Q9H0Z5, Q9H109 | Gene names | ID1, ID | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
HES6_MOUSE
|
||||||
| NC score | 0.087281 (rank : 70) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.087053 (rank : 71) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
HES2_HUMAN
|
||||||
| NC score | 0.084665 (rank : 72) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES3_MOUSE
|
||||||
| NC score | 0.081597 (rank : 73) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.080931 (rank : 74) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_HUMAN
|
||||||
| NC score | 0.080252 (rank : 75) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.080152 (rank : 76) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
HES7_HUMAN
|
||||||
| NC score | 0.077041 (rank : 77) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| NC score | 0.076462 (rank : 78) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.074460 (rank : 79) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
HAND2_HUMAN
|
||||||
| NC score | 0.074297 (rank : 80) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| NC score | 0.074297 (rank : 81) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
HAND1_HUMAN
|
||||||
| NC score | 0.068336 (rank : 82) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.067571 (rank : 83) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.066840 (rank : 84) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.066095 (rank : 85) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.065831 (rank : 86) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HAND1_MOUSE
|
||||||
| NC score | 0.065468 (rank : 87) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.065402 (rank : 88) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.065290 (rank : 89) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.064860 (rank : 90) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
TWST1_HUMAN
|
||||||
| NC score | 0.064722 (rank : 91) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| NC score | 0.064645 (rank : 92) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.064308 (rank : 93) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.063902 (rank : 94) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
TWST2_MOUSE
|
||||||
| NC score | 0.063821 (rank : 95) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_HUMAN
|
||||||
| NC score | 0.063723 (rank : 96) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.063645 (rank : 97) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.063529 (rank : 98) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
PTF1A_HUMAN
|
||||||
| NC score | 0.063103 (rank : 99) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
LYL1_HUMAN
|
||||||
| NC score | 0.061719 (rank : 100) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
PTF1A_MOUSE
|
||||||
| NC score | 0.061716 (rank : 101) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.060957 (rank : 102) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.059552 (rank : 103) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
LYL1_MOUSE
|
||||||
| NC score | 0.059426 (rank : 104) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.058502 (rank : 105) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
TAL2_MOUSE
|
||||||
| NC score | 0.058048 (rank : 106) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
PHAR2_HUMAN
|
||||||
| NC score | 0.057834 (rank : 107) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.056864 (rank : 108) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.056464 (rank : 109) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.055194 (rank : 110) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.054624 (rank : 111) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
ASCL1_HUMAN
|
||||||
| NC score | 0.053840 (rank : 112) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
MYF5_HUMAN
|
||||||
| NC score | 0.053839 (rank : 113) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.053711 (rank : 114) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYF5_MOUSE
|
||||||
| NC score | 0.053610 (rank : 115) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.053426 (rank : 116) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
SCX_MOUSE
|
||||||
| NC score | 0.052729 (rank : 117) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |

|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.052586 (rank : 118) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
TAL2_HUMAN
|
||||||
| NC score | 0.050996 (rank : 119) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
ASCL2_MOUSE
|
||||||
| NC score | 0.050823 (rank : 120) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
ASCL1_MOUSE
|
||||||
| NC score | 0.050592 (rank : 121) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.050227 (rank : 122) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
ITF2_MOUSE
|
||||||
| NC score | 0.048880 (rank : 123) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
NGN1_MOUSE
|
||||||
| NC score | 0.047019 (rank : 124) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.046095 (rank : 125) | θ value | 0.0736092 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.045910 (rank : 126) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.039850 (rank : 127) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.036859 (rank : 128) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
XLR_MOUSE
|
||||||
| NC score | 0.035703 (rank : 129) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05531 | Gene names | Xlr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked lymphocyte-regulated protein PM1. | |||||
|
TRAK2_HUMAN
|
||||||
| NC score | 0.035484 (rank : 130) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |

|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
HOME2_HUMAN
|
||||||
| NC score | 0.035324 (rank : 131) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NSB8, O95269, O95349, Q9NSB6, Q9NSB7, Q9UNT7 | Gene names | HOMER2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.034192 (rank : 132) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.033988 (rank : 133) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
CDC37_MOUSE
|
||||||
| NC score | 0.033953 (rank : 134) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.033409 (rank : 135) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
CV019_HUMAN
|
||||||
| NC score | 0.033079 (rank : 136) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13769, O60839, Q9UPZ5 | Gene names | C22orf19, KIAA0983 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C22orf19 (NF2/meningioma region protein pK1.3) (Placental protein 39.2). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.030947 (rank : 137) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.030389 (rank : 138) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
TULP1_MOUSE
|
||||||
| NC score | 0.030106 (rank : 139) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.029704 (rank : 140) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.029658 (rank : 141) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.029613 (rank : 142) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.028139 (rank : 143) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
CCD11_MOUSE
|
||||||
| NC score | 0.028002 (rank : 144) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D439 | Gene names | Ccdc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.027522 (rank : 145) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CCD27_MOUSE
|
||||||
| NC score | 0.026874 (rank : 146) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.026236 (rank : 147) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.025962 (rank : 148) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.025520 (rank : 149) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
MFAP1_MOUSE
|
||||||
| NC score | 0.025348 (rank : 150) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CQU1, Q3TU29, Q8CCL1, Q9CSJ5 | Gene names | Mfap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microfibrillar-associated protein 1. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.024434 (rank : 151) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.022960 (rank : 152) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.022892 (rank : 153) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
PLXD1_HUMAN
|
||||||
| NC score | 0.018641 (rank : 154) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
ERBB3_HUMAN
|
||||||
| NC score | 0.015093 (rank : 155) | θ value | 0.163984 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
SAS10_HUMAN
|
||||||
| NC score | 0.013272 (rank : 156) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQZ2, Q6FI82 | Gene names | CRLZ1, SAS10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.013041 (rank : 157) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
ERBB3_MOUSE
|
||||||
| NC score | 0.012421 (rank : 158) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.011986 (rank : 159) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.011732 (rank : 160) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
LAMB2_HUMAN
|
||||||
| NC score | 0.011064 (rank : 161) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
CLK1_HUMAN
|
||||||
| NC score | 0.004127 (rank : 162) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49759 | Gene names | CLK1, CLK | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase CLK1 (EC 2.7.12.1) (CDC-like kinase 1). | |||||
|
PCDGM_HUMAN
|
||||||
| NC score | 0.001649 (rank : 163) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5F6, Q9Y5C2 | Gene names | PCDHGC5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C5 precursor (PCDH-gamma-C5). | |||||