Please be patient as the page loads
|
NPAS1_MOUSE
|
||||||
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NPAS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994444 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 2.00348e-116 (rank : 3) | NC score | 0.968990 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 1.69605e-115 (rank : 4) | NC score | 0.971139 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 1.5519e-76 (rank : 5) | NC score | 0.952717 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 5.8972e-76 (rank : 6) | NC score | 0.954256 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 4.99229e-75 (rank : 7) | NC score | 0.947077 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM1_HUMAN
|
||||||
| θ value | 6.52014e-75 (rank : 8) | NC score | 0.947809 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 1.23823e-49 (rank : 9) | NC score | 0.900850 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 4.70527e-49 (rank : 10) | NC score | 0.901675 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 8.02596e-49 (rank : 11) | NC score | 0.909256 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 1.04822e-48 (rank : 12) | NC score | 0.906407 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 13) | NC score | 0.747095 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 14) | NC score | 0.744462 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 15) | NC score | 0.801844 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 16) | NC score | 0.798201 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 17) | NC score | 0.704600 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 18) | NC score | 0.706412 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 19) | NC score | 0.687367 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 20) | NC score | 0.685983 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 21) | NC score | 0.662743 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 22) | NC score | 0.650031 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 23) | NC score | 0.611396 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 24) | NC score | 0.608617 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 25) | NC score | 0.662502 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 26) | NC score | 0.544182 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 27) | NC score | 0.544865 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 28) | NC score | 0.673038 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 29) | NC score | 0.673607 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 30) | NC score | 0.664367 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 31) | NC score | 0.537310 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 32) | NC score | 0.501054 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.005199 (rank : 79) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.029516 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.024472 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
LYL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.023966 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.021619 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.016782 (rank : 66) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.018578 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TAL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.025758 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
TAL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.023402 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.022097 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.007456 (rank : 75) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.015554 (rank : 68) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.015903 (rank : 67) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.014538 (rank : 69) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.004500 (rank : 80) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
GCH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.016783 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30793, Q9Y4I8 | Gene names | GCH1, DYT5, GCH | |||
|
Domain Architecture |

|
|||||
| Description | GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.009932 (rank : 72) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
ABCBA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.003069 (rank : 81) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRK6, Q13040, Q9H3V0 | Gene names | ABCB10 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family B member 10, mitochondrial precursor (ATP-binding cassette transporter 10) (ABC transporter 10 protein) (Mitochondrial ATP-binding cassette 2) (M-ABC2). | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | -0.000017 (rank : 84) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
MYBB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.009567 (rank : 73) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10244 | Gene names | MYBL2, BMYB | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.008788 (rank : 74) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.014430 (rank : 70) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.006400 (rank : 77) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
KLF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | -0.001470 (rank : 85) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60843 | Gene names | Klf2, Lklf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 2 (Lung krueppel-like factor). | |||||
|
LYL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.018596 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.020882 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
LMO6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.002203 (rank : 83) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |
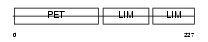
|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.005296 (rank : 78) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.010746 (rank : 71) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
STX18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.007095 (rank : 76) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.002678 (rank : 82) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.052823 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.075789 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.079145 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.085313 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.083912 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.058166 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.059224 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.055104 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.055194 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
PER1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.163680 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.161573 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.168204 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.152180 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PER3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.160392 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.182062 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.060532 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.056068 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.055376 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.055388 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.052457 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.052433 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.051055 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.994444 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.971139 (rank : 3) | θ value | 1.69605e-115 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.968990 (rank : 4) | θ value | 2.00348e-116 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.954256 (rank : 5) | θ value | 5.8972e-76 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.952717 (rank : 6) | θ value | 1.5519e-76 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.947809 (rank : 7) | θ value | 6.52014e-75 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.947077 (rank : 8) | θ value | 4.99229e-75 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.909256 (rank : 9) | θ value | 8.02596e-49 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.906407 (rank : 10) | θ value | 1.04822e-48 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.901675 (rank : 11) | θ value | 4.70527e-49 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.900850 (rank : 12) | θ value | 1.23823e-49 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.801844 (rank : 13) | θ value | 1.058e-16 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.798201 (rank : 14) | θ value | 1.80466e-16 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.747095 (rank : 15) | θ value | 1.6852e-22 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.744462 (rank : 16) | θ value | 8.36355e-22 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.706412 (rank : 17) | θ value | 6.41864e-14 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.704600 (rank : 18) | θ value | 2.20605e-14 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.687367 (rank : 19) | θ value | 2.43908e-13 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.685983 (rank : 20) | θ value | 4.16044e-13 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.673607 (rank : 21) | θ value | 3.64472e-09 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.673038 (rank : 22) | θ value | 9.59137e-10 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.664367 (rank : 23) | θ value | 1.38499e-08 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.662743 (rank : 24) | θ value | 1.2105e-12 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.662502 (rank : 25) | θ value | 3.89403e-11 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.650031 (rank : 26) | θ value | 3.52202e-12 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.611396 (rank : 27) | θ value | 7.84624e-12 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.608617 (rank : 28) | θ value | 1.33837e-11 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.544865 (rank : 29) | θ value | 2.52405e-10 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.544182 (rank : 30) | θ value | 8.67504e-11 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.537310 (rank : 31) | θ value | 6.87365e-08 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.501054 (rank : 32) | θ value | 6.43352e-06 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.182062 (rank : 33) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.168204 (rank : 34) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.163680 (rank : 35) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.161573 (rank : 36) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.160392 (rank : 37) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.152180 (rank : 38) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.085313 (rank : 39) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.083912 (rank : 40) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.079145 (rank : 41) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.075789 (rank : 42) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.060532 (rank : 43) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.059224 (rank : 44) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.058166 (rank : 45) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.056068 (rank : 46) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.055388 (rank : 47) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.055376 (rank : 48) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.055194 (rank : 49) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.055104 (rank : 50) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.052823 (rank : 51) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.052457 (rank : 52) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.052433 (rank : 53) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.051055 (rank : 54) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.029516 (rank : 55) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
TAL1_HUMAN
|
||||||
| NC score | 0.025758 (rank : 56) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.024472 (rank : 57) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
LYL1_HUMAN
|
||||||
| NC score | 0.023966 (rank : 58) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
TAL1_MOUSE
|
||||||
| NC score | 0.023402 (rank : 59) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.022097 (rank : 60) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.021619 (rank : 61) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.020882 (rank : 62) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
LYL1_MOUSE
|
||||||
| NC score | 0.018596 (rank : 63) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.018578 (rank : 64) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
GCH1_HUMAN
|
||||||
| NC score | 0.016783 (rank : 65) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30793, Q9Y4I8 | Gene names | GCH1, DYT5, GCH | |||
|
Domain Architecture |

|
|||||
| Description | GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.016782 (rank : 66) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.015903 (rank : 67) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.015554 (rank : 68) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.014538 (rank : 69) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.014430 (rank : 70) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.010746 (rank : 71) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.009932 (rank : 72) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
MYBB_HUMAN
|
||||||
| NC score | 0.009567 (rank : 73) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10244 | Gene names | MYBL2, BMYB | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.008788 (rank : 74) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.007456 (rank : 75) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
STX18_HUMAN
|
||||||
| NC score | 0.007095 (rank : 76) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.006400 (rank : 77) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.005296 (rank : 78) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | 0.005199 (rank : 79) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.004500 (rank : 80) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
ABCBA_HUMAN
|
||||||
| NC score | 0.003069 (rank : 81) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRK6, Q13040, Q9H3V0 | Gene names | ABCB10 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family B member 10, mitochondrial precursor (ATP-binding cassette transporter 10) (ABC transporter 10 protein) (Mitochondrial ATP-binding cassette 2) (M-ABC2). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.002678 (rank : 82) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
LMO6_HUMAN
|
||||||
| NC score | 0.002203 (rank : 83) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
EGR4_HUMAN
|
||||||
| NC score | -0.000017 (rank : 84) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
KLF2_MOUSE
|
||||||
| NC score | -0.001470 (rank : 85) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60843 | Gene names | Klf2, Lklf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 2 (Lung krueppel-like factor). | |||||