Please be patient as the page loads
|
NPAS3_HUMAN
|
||||||
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NPAS3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996604 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | 3.41741e-116 (rank : 3) | NC score | 0.970903 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 1.69605e-115 (rank : 4) | NC score | 0.971139 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 2.0926e-81 (rank : 5) | NC score | 0.946602 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 4.66183e-81 (rank : 6) | NC score | 0.944354 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| θ value | 3.34088e-79 (rank : 7) | NC score | 0.938817 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 2.1655e-78 (rank : 8) | NC score | 0.938104 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 9) | NC score | 0.905775 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 1.36585e-56 (rank : 10) | NC score | 0.899765 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 1.36585e-56 (rank : 11) | NC score | 0.907626 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 3.974e-56 (rank : 12) | NC score | 0.900651 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 2.77775e-25 (rank : 13) | NC score | 0.815377 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 1.80048e-24 (rank : 14) | NC score | 0.810915 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 15) | NC score | 0.754424 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 9.87957e-23 (rank : 16) | NC score | 0.751635 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 17) | NC score | 0.694427 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 18) | NC score | 0.719957 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 19) | NC score | 0.717795 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 20) | NC score | 0.682043 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 21) | NC score | 0.702335 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 22) | NC score | 0.700658 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 23) | NC score | 0.690279 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 24) | NC score | 0.682649 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 25) | NC score | 0.596109 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 26) | NC score | 0.598693 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 27) | NC score | 0.651338 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 28) | NC score | 0.638330 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 29) | NC score | 0.527663 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 30) | NC score | 0.526521 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 31) | NC score | 0.521640 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 32) | NC score | 0.486245 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 33) | NC score | 0.057558 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 34) | NC score | 0.049798 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 35) | NC score | 0.042869 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 36) | NC score | 0.048342 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
ZN483_HUMAN
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | -0.001324 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TF39 | Gene names | ZNF483, KIAA1962 | |||
|
Domain Architecture |
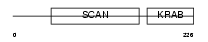
|
|||||
| Description | Zinc finger protein 483. | |||||
|
MYCD_HUMAN
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.030399 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.022269 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.034290 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LPIN2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.024297 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
MIA2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.044942 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
JHD2A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.021146 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PCM1, Q2MJQ6, Q3TKW8, Q3UML3, Q6ZQ57, Q8K2J6, Q8K2K4, Q8R350 | Gene names | Jmjd1a, Jhdm2a, Kiaa0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.020345 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PZRN4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.012703 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
NC6IP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.043265 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RS0, Q5GH23, Q8TDG9, Q96QU3, Q9H5V3 | Gene names | NCOA6IP, PIMT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT) (CLL-associated antigen KW-2) (Hepatocellular carcinoma-associated antigen 137) (HCA137). | |||||
|
AFTIN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.018868 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80WT5, Q5SSE6, Q99KJ1 | Gene names | Aftph, Afth | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aftiphilin. | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.025854 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.027817 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.022055 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.026875 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.011316 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SOCS6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.010442 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLY0, Q9D2A0 | Gene names | Socs6, Cis4, Cish4, Socs4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 6 (SOCS-6) (Suppressor of cytokine signaling 4) (SOCS-4) (Cytokine-inducible SH2 protein 4) (CIS-4). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.021030 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.023881 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.038381 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.101841 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.100058 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | -0.002488 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MCES_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.020939 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
TBX2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.004110 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.013762 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.007790 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
GCP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.019095 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RT8, Q96PY8 | Gene names | TUBGCP5, GCP5, KIAA1899 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 5 (GCP-5). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.091564 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.095009 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
NC6IP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.030036 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
OS9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.028693 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.007677 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.171833 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
TIAM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.008526 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
TSP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.006024 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07996, Q15667 | Gene names | THBS1, TSP, TSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
TSP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.005592 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49746, Q8WV34 | Gene names | THBS3, TSP3 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-3 precursor. | |||||
|
DSG1C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.003186 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TSF0, Q7TQ61 | Gene names | Dsg1c, Dsg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 gamma precursor (Dsg1-gamma) (Desmoglein-6). | |||||
|
MMS19_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.012715 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D071, Q925N8, Q9CWX3 | Gene names | Mms19l, Mms19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MMS19-like protein (MET18 homolog). | |||||
|
TSP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.005337 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
ZBT40_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | -0.001752 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.009590 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
FA35A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.008680 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UEN2 | Gene names | Fam35a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM35A. | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.001274 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.005038 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.061048 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.062481 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.063754 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.058510 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.058502 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
PER1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.173251 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.176599 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.158886 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PER3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.166333 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.189462 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.063824 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.062120 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.059718 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.059686 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.058265 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.058238 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.056283 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.052539 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.996604 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.971139 (rank : 3) | θ value | 1.69605e-115 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.970903 (rank : 4) | θ value | 3.41741e-116 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.946602 (rank : 5) | θ value | 2.0926e-81 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.944354 (rank : 6) | θ value | 4.66183e-81 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.938817 (rank : 7) | θ value | 3.34088e-79 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.938104 (rank : 8) | θ value | 2.1655e-78 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.907626 (rank : 9) | θ value | 1.36585e-56 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.905775 (rank : 10) | θ value | 8.55514e-59 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.900651 (rank : 11) | θ value | 3.974e-56 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.899765 (rank : 12) | θ value | 1.36585e-56 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.815377 (rank : 13) | θ value | 2.77775e-25 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.810915 (rank : 14) | θ value | 1.80048e-24 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.754424 (rank : 15) | θ value | 1.5242e-23 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.751635 (rank : 16) | θ value | 9.87957e-23 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.719957 (rank : 17) | θ value | 7.32683e-18 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.717795 (rank : 18) | θ value | 9.56915e-18 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.702335 (rank : 19) | θ value | 4.02038e-16 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.700658 (rank : 20) | θ value | 8.95645e-16 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.694427 (rank : 21) | θ value | 5.60996e-18 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.690279 (rank : 22) | θ value | 1.86753e-13 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.682649 (rank : 23) | θ value | 5.43371e-13 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.682043 (rank : 24) | θ value | 1.63225e-17 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.651338 (rank : 25) | θ value | 7.84624e-12 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.638330 (rank : 26) | θ value | 1.02475e-11 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.598693 (rank : 27) | θ value | 2.69671e-12 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.596109 (rank : 28) | θ value | 2.69671e-12 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.527663 (rank : 29) | θ value | 6.21693e-09 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.526521 (rank : 30) | θ value | 1.06045e-08 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.521640 (rank : 31) | θ value | 7.59969e-07 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.486245 (rank : 32) | θ value | 6.43352e-06 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.189462 (rank : 33) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.176599 (rank : 34) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.173251 (rank : 35) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.171833 (rank : 36) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.166333 (rank : 37) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.158886 (rank : 38) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.101841 (rank : 39) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.100058 (rank : 40) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.095009 (rank : 41) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.091564 (rank : 42) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.063824 (rank : 43) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.063754 (rank : 44) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.062481 (rank : 45) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.062120 (rank : 46) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.061048 (rank : 47) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.059718 (rank : 48) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.059686 (rank : 49) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.058510 (rank : 50) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.058502 (rank : 51) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.058265 (rank : 52) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.058238 (rank : 53) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.057558 (rank : 54) | θ value | 0.00390308 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.056283 (rank : 55) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.052539 (rank : 56) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.049798 (rank : 57) | θ value | 0.00390308 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.048342 (rank : 58) | θ value | 0.0330416 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MIA2_HUMAN
|
||||||
| NC score | 0.044942 (rank : 59) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
NC6IP_HUMAN
|
||||||
| NC score | 0.043265 (rank : 60) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RS0, Q5GH23, Q8TDG9, Q96QU3, Q9H5V3 | Gene names | NCOA6IP, PIMT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT) (CLL-associated antigen KW-2) (Hepatocellular carcinoma-associated antigen 137) (HCA137). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.042869 (rank : 61) | θ value | 0.00869519 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.038381 (rank : 62) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.034290 (rank : 63) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYCD_HUMAN
|
||||||
| NC score | 0.030399 (rank : 64) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
NC6IP_MOUSE
|
||||||
| NC score | 0.030036 (rank : 65) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
OS9_HUMAN
|
||||||
| NC score | 0.028693 (rank : 66) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.027817 (rank : 67) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.026875 (rank : 68) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.025854 (rank : 69) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
LPIN2_HUMAN
|
||||||
| NC score | 0.024297 (rank : 70) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.023881 (rank : 71) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.022269 (rank : 72) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.022055 (rank : 73) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
JHD2A_MOUSE
|
||||||
| NC score | 0.021146 (rank : 74) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PCM1, Q2MJQ6, Q3TKW8, Q3UML3, Q6ZQ57, Q8K2J6, Q8K2K4, Q8R350 | Gene names | Jmjd1a, Jhdm2a, Kiaa0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.021030 (rank : 75) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
MCES_HUMAN
|
||||||
| NC score | 0.020939 (rank : 76) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.020345 (rank : 77) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
GCP5_HUMAN
|
||||||
| NC score | 0.019095 (rank : 78) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RT8, Q96PY8 | Gene names | TUBGCP5, GCP5, KIAA1899 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 5 (GCP-5). | |||||
|
AFTIN_MOUSE
|
||||||
| NC score | 0.018868 (rank : 79) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80WT5, Q5SSE6, Q99KJ1 | Gene names | Aftph, Afth | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aftiphilin. | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.013762 (rank : 80) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
MMS19_MOUSE
|
||||||
| NC score | 0.012715 (rank : 81) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D071, Q925N8, Q9CWX3 | Gene names | Mms19l, Mms19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MMS19-like protein (MET18 homolog). | |||||
|
PZRN4_HUMAN
|
||||||
| NC score | 0.012703 (rank : 82) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.011316 (rank : 83) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SOCS6_MOUSE
|
||||||
| NC score | 0.010442 (rank : 84) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLY0, Q9D2A0 | Gene names | Socs6, Cis4, Cish4, Socs4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 6 (SOCS-6) (Suppressor of cytokine signaling 4) (SOCS-4) (Cytokine-inducible SH2 protein 4) (CIS-4). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.009590 (rank : 85) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
FA35A_MOUSE
|
||||||
| NC score | 0.008680 (rank : 86) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UEN2 | Gene names | Fam35a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM35A. | |||||
|
TIAM1_MOUSE
|
||||||
| NC score | 0.008526 (rank : 87) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.007790 (rank : 88) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.007677 (rank : 89) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
TSP1_HUMAN
|
||||||
| NC score | 0.006024 (rank : 90) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07996, Q15667 | Gene names | THBS1, TSP, TSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
TSP3_HUMAN
|
||||||
| NC score | 0.005592 (rank : 91) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49746, Q8WV34 | Gene names | THBS3, TSP3 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-3 precursor. | |||||
|
TSP1_MOUSE
|
||||||
| NC score | 0.005337 (rank : 92) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.005038 (rank : 93) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
TBX2_HUMAN
|
||||||
| NC score | 0.004110 (rank : 94) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
DSG1C_MOUSE
|
||||||
| NC score | 0.003186 (rank : 95) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TSF0, Q7TQ61 | Gene names | Dsg1c, Dsg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 gamma precursor (Dsg1-gamma) (Desmoglein-6). | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.001274 (rank : 96) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
ZN483_HUMAN
|
||||||
| NC score | -0.001324 (rank : 97) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TF39 | Gene names | ZNF483, KIAA1962 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 483. | |||||
|
ZBT40_HUMAN
|
||||||
| NC score | -0.001752 (rank : 98) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | -0.002488 (rank : 99) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||