Please be patient as the page loads
|
LPIN2_HUMAN
|
||||||
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LPIN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.971310 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14693 | Gene names | LPIN1, KIAA0188 | |||
|
Domain Architecture |
|
|||||
| Description | Lipin-1. | |||||
|
LPIN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.961213 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91ZP3, Q9CQI2, Q9JLG6 | Gene names | Lpin1, Fld | |||
|
Domain Architecture |
|
|||||
| Description | Lipin-1 (Fatty liver dystrophy protein). | |||||
|
LPIN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
LPIN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.985329 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PI5, Q8C357, Q8C7I8, Q8CC85, Q8CHR7 | Gene names | Lpin2 | |||
|
Domain Architecture |
|
|||||
| Description | Lipin-2. | |||||
|
LPIN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.965992 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
LPIN3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.968404 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |
|
|||||
| Description | Lipin-3. | |||||
|
PMS2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.077323 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 8) | NC score | 0.054859 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.007443 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 10) | NC score | 0.065710 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.049691 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.043274 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.052282 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.022739 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.050867 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
RP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.034607 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |
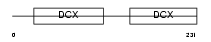
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
BIN1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.050136 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.051663 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
TTK_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.006654 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33981 | Gene names | TTK, MPS1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (Phosphotyrosine picked threonine-protein kinase) (PYT). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.041827 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BASP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.047053 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.023987 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.027582 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.022440 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
B3A3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.015425 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.032619 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.024297 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.025183 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.037851 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.027110 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.035786 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
AF1Q_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.082300 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13015 | Gene names | MLLT11, AF1Q | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein AF1q. | |||||
|
AF1Q_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.082133 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97783 | Gene names | Mllt11, Af1q | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein AF1q. | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.022746 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.046350 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
REPS2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.032115 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XA6 | Gene names | Reps2, Pob1 | |||
|
Domain Architecture |
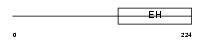
|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.018438 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
MLH1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.031634 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK91, Q62454 | Gene names | Mlh1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
SAC3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.021702 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92562, Q53H49, Q5TCS6 | Gene names | SAC3, KIAA0274 | |||
|
Domain Architecture |
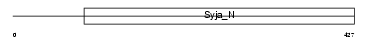
|
|||||
| Description | SAC domain-containing protein 3. | |||||
|
SPRE2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.024020 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z698, Q2NKX6 | Gene names | SPRED2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.030253 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.010492 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.030398 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.022961 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SPRE2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.023488 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q924S7 | Gene names | Spred2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.021247 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.012532 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.014303 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.005401 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.022886 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
BAT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.020154 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
CI040_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.074224 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IXQ3, Q9NWD3 | Gene names | C9orf40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf40. | |||||
|
GLE1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.012416 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
PALB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.020417 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YC2, Q8N7Y6, Q8ND31, Q9H6W1 | Gene names | PALB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.036362 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.020985 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.011432 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.022512 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
CLMN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.010143 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.036059 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.014696 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NOB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.065651 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULX3, Q7L6B7, Q7M4M4, Q7Z4B5, Q9NWB0 | Gene names | NOB1, ART4, NOB1P, PSMD8BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein NOB1 (Protein ART-4) (Phosphorylation regulatory protein HP-10). | |||||
|
PP4R1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.029523 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TF05, Q99774, Q9UNQ7 | Gene names | PPP4R1, MEG1, PP4R1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.036162 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
SAPS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.014108 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
SP110_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.015437 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.019749 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
ZN206_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | -0.000967 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96SZ4 | Gene names | ZNF206, ZSCAN10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 206 (Zinc finger and SCAN domain-containing protein 10). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.023196 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.014964 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.015539 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
HM2L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.018331 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
M3K12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.000772 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.020973 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MRP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.020007 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
PP4R1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.029884 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.006920 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.015420 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.039770 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.012176 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.019461 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CEBPZ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.014096 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53569, Q3TXW5, Q3TYA8, Q3UWQ7 | Gene names | Cebpz, Cbf2, Cebpa-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein zeta (CCAAT-box-binding transcription factor) (CCAAT-binding factor) (CBF). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.014593 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
IFI3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.010020 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35368 | Gene names | Ifi203 | |||
|
Domain Architecture |
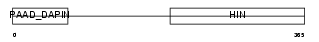
|
|||||
| Description | Interferon-activable protein 203 (Ifi-203) (Interferon-inducible protein p203). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.029344 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.014762 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NOB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.071742 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BW10, Q7TPR3, Q9CRD7 | Gene names | Nob1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein NOB1. | |||||
|
ZF276_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.000027 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.019070 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
FIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.012800 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.017908 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.021952 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.008959 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.015172 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PHF22_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.013555 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
RFFL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.020290 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
RGAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.005340 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.021620 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TMED8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.013276 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PL24, Q9P1V9 | Gene names | TMED8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TMED8. | |||||
|
WIPI3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.018368 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5MNZ6, O95328, Q6IBN2 | Gene names | WDR45L, WIPI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat domain phosphoinositide-interacting protein 3 (WIPI-3) (WD repeat protein 45-like) (WDR45-like protein) (WIPI49-like protein). | |||||
|
ZNF18_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | -0.001512 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810A1, Q9D972 | Gene names | Znf18, Zfp535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 18 (Zinc finger protein 535). | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.012638 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.033101 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ALS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.016133 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
BMP3B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.003578 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97737 | Gene names | Gdf10, Bmp3b | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3b precursor (BMP-3b) (Growth/differentiation factor 10) (GDF-10). | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.020746 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
DMC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.009925 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14565, Q99498, Q9UH11 | Gene names | DMC1, DMC1H, LIM15 | |||
|
Domain Architecture |

|
|||||
| Description | Meiotic recombination protein DMC1/LIM15 homolog. | |||||
|
DMC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.009918 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61880 | Gene names | Dmc1, Dmc1h, Lim15 | |||
|
Domain Architecture |

|
|||||
| Description | Meiotic recombination protein DMC1/LIM15 homolog. | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.007223 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
HN1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.014351 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H910, Q6EIC7 | Gene names | C16orf34, HN1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematological and neurological expressed 1-like protein (HN1-like protein). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.002742 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
MAGD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.008392 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
SAPS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.011653 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
SRP72_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.011742 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O76094 | Gene names | SRP72 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 72 kDa protein (SRP72). | |||||
|
TEP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.015476 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97499 | Gene names | Tep1, Tp1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 homolog). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.021947 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.026452 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
LPIN2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
LPIN2_MOUSE
|
||||||
| NC score | 0.985329 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PI5, Q8C357, Q8C7I8, Q8CC85, Q8CHR7 | Gene names | Lpin2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-2. | |||||
|
LPIN1_HUMAN
|
||||||
| NC score | 0.971310 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14693 | Gene names | LPIN1, KIAA0188 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-1. | |||||
|
LPIN3_MOUSE
|
||||||
| NC score | 0.968404 (rank : 4) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-3. | |||||
|
LPIN3_HUMAN
|
||||||
| NC score | 0.965992 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
LPIN1_MOUSE
|
||||||
| NC score | 0.961213 (rank : 6) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91ZP3, Q9CQI2, Q9JLG6 | Gene names | Lpin1, Fld | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-1 (Fatty liver dystrophy protein). | |||||
|
AF1Q_HUMAN
|
||||||
| NC score | 0.082300 (rank : 7) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13015 | Gene names | MLLT11, AF1Q | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein AF1q. | |||||
|
AF1Q_MOUSE
|
||||||
| NC score | 0.082133 (rank : 8) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97783 | Gene names | Mllt11, Af1q | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein AF1q. | |||||
|
PMS2_HUMAN
|
||||||
| NC score | 0.077323 (rank : 9) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
CI040_HUMAN
|
||||||
| NC score | 0.074224 (rank : 10) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IXQ3, Q9NWD3 | Gene names | C9orf40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf40. | |||||
|
NOB1_MOUSE
|
||||||
| NC score | 0.071742 (rank : 11) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BW10, Q7TPR3, Q9CRD7 | Gene names | Nob1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein NOB1. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.065710 (rank : 12) | θ value | 0.0330416 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NOB1_HUMAN
|
||||||
| NC score | 0.065651 (rank : 13) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULX3, Q7L6B7, Q7M4M4, Q7Z4B5, Q9NWB0 | Gene names | NOB1, ART4, NOB1P, PSMD8BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein NOB1 (Protein ART-4) (Phosphorylation regulatory protein HP-10). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.054859 (rank : 14) | θ value | 0.0193708 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.052282 (rank : 15) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.051663 (rank : 16) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.050867 (rank : 17) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
BIN1_MOUSE
|
||||||
| NC score | 0.050136 (rank : 18) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.049691 (rank : 19) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.047053 (rank : 20) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.046350 (rank : 21) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.043274 (rank : 22) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.041827 (rank : 23) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.039770 (rank : 24) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.037851 (rank : 25) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.036362 (rank : 26) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.036162 (rank : 27) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.036059 (rank : 28) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.035786 (rank : 29) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RP1_MOUSE
|
||||||
| NC score | 0.034607 (rank : 30) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.033101 (rank : 31) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.032619 (rank : 32) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
REPS2_MOUSE
|
||||||
| NC score | 0.032115 (rank : 33) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XA6 | Gene names | Reps2, Pob1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
MLH1_MOUSE
|
||||||
| NC score | 0.031634 (rank : 34) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK91, Q62454 | Gene names | Mlh1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.030398 (rank : 35) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.030253 (rank : 36) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PP4R1_MOUSE
|
||||||
| NC score | 0.029884 (rank : 37) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
PP4R1_HUMAN
|
||||||
| NC score | 0.029523 (rank : 38) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TF05, Q99774, Q9UNQ7 | Gene names | PPP4R1, MEG1, PP4R1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.029344 (rank : 39) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.027582 (rank : 40) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.027110 (rank : 41) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.026452 (rank : 42) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.025183 (rank : 43) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.024297 (rank : 44) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
SPRE2_HUMAN
|
||||||
| NC score | 0.024020 (rank : 45) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z698, Q2NKX6 | Gene names | SPRED2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.023987 (rank : 46) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
SPRE2_MOUSE
|
||||||
| NC score | 0.023488 (rank : 47) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q924S7 | Gene names | Spred2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.023196 (rank : 48) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.022961 (rank : 49) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.022886 (rank : 50) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.022746 (rank : 51) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.022739 (rank : 52) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.022512 (rank : 53) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.022440 (rank : 54) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.021952 (rank : 55) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.021947 (rank : 56) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
SAC3_HUMAN
|
||||||
| NC score | 0.021702 (rank : 57) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92562, Q53H49, Q5TCS6 | Gene names | SAC3, KIAA0274 | |||
|
Domain Architecture |

|
|||||
| Description | SAC domain-containing protein 3. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.021620 (rank : 58) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.021247 (rank : 59) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.020985 (rank : 60) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.020973 (rank : 61) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.020746 (rank : 62) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
PALB2_HUMAN
|
||||||
| NC score | 0.020417 (rank : 63) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YC2, Q8N7Y6, Q8ND31, Q9H6W1 | Gene names | PALB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
RFFL_HUMAN
|
||||||
| NC score | 0.020290 (rank : 64) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.020154 (rank : 65) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.020007 (rank : 66) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.019749 (rank : 67) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.019461 (rank : 68) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.019070 (rank : 69) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.018438 (rank : 70) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
WIPI3_HUMAN
|
||||||
| NC score | 0.018368 (rank : 71) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5MNZ6, O95328, Q6IBN2 | Gene names | WDR45L, WIPI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat domain phosphoinositide-interacting protein 3 (WIPI-3) (WD repeat protein 45-like) (WDR45-like protein) (WIPI49-like protein). | |||||
|
HM2L1_HUMAN
|
||||||
| NC score | 0.018331 (rank : 72) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.017908 (rank : 73) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.016133 (rank : 74) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.015539 (rank : 75) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
TEP1_MOUSE
|
||||||
| NC score | 0.015476 (rank : 76) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97499 | Gene names | Tep1, Tp1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 homolog). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.015437 (rank : 77) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
B3A3_MOUSE
|
||||||
| NC score | 0.015425 (rank : 78) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.015420 (rank : 79) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.015172 (rank : 80) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.014964 (rank : 81) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.014762 (rank : 82) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.014696 (rank : 83) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.014593 (rank : 84) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
HN1L_HUMAN
|
||||||
| NC score | 0.014351 (rank : 85) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H910, Q6EIC7 | Gene names | C16orf34, HN1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematological and neurological expressed 1-like protein (HN1-like protein). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.014303 (rank : 86) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
SAPS3_HUMAN
|
||||||
| NC score | 0.014108 (rank : 87) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
CEBPZ_MOUSE
|
||||||
| NC score | 0.014096 (rank : 88) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53569, Q3TXW5, Q3TYA8, Q3UWQ7 | Gene names | Cebpz, Cbf2, Cebpa-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein zeta (CCAAT-box-binding transcription factor) (CCAAT-binding factor) (CBF). | |||||
|
PHF22_MOUSE
|
||||||
| NC score | 0.013555 (rank : 89) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
TMED8_HUMAN
|
||||||
| NC score | 0.013276 (rank : 90) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PL24, Q9P1V9 | Gene names | TMED8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TMED8. | |||||
|
FIP1_MOUSE
|
||||||
| NC score | 0.012800 (rank : 91) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.012638 (rank : 92) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.012532 (rank : 93) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
GLE1_MOUSE
|
||||||
| NC score | 0.012416 (rank : 94) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.012176 (rank : 95) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
SRP72_HUMAN
|
||||||
| NC score | 0.011742 (rank : 96) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O76094 | Gene names | SRP72 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 72 kDa protein (SRP72). | |||||
|
SAPS3_MOUSE
|
||||||
| NC score | 0.011653 (rank : 97) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.011432 (rank : 98) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.010492 (rank : 99) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.010143 (rank : 100) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
IFI3_MOUSE
|
||||||
| NC score | 0.010020 (rank : 101) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35368 | Gene names | Ifi203 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 203 (Ifi-203) (Interferon-inducible protein p203). | |||||
|
DMC1_HUMAN
|
||||||
| NC score | 0.009925 (rank : 102) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14565, Q99498, Q9UH11 | Gene names | DMC1, DMC1H, LIM15 | |||
|
Domain Architecture |

|
|||||
| Description | Meiotic recombination protein DMC1/LIM15 homolog. | |||||
|
DMC1_MOUSE
|
||||||
| NC score | 0.009918 (rank : 103) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61880 | Gene names | Dmc1, Dmc1h, Lim15 | |||
|
Domain Architecture |

|
|||||
| Description | Meiotic recombination protein DMC1/LIM15 homolog. | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.008959 (rank : 104) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
MAGD2_HUMAN
|
||||||
| NC score | 0.008392 (rank : 105) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.007443 (rank : 106) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.007223 (rank : 107) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.006920 (rank : 108) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
TTK_HUMAN
|
||||||
| NC score | 0.006654 (rank : 109) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33981 | Gene names | TTK, MPS1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (Phosphotyrosine picked threonine-protein kinase) (PYT). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.005401 (rank : 110) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
RGAP1_MOUSE
|
||||||
| NC score | 0.005340 (rank : 111) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
BMP3B_MOUSE
|
||||||
| NC score | 0.003578 (rank : 112) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97737 | Gene names | Gdf10, Bmp3b | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3b precursor (BMP-3b) (Growth/differentiation factor 10) (GDF-10). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.002742 (rank : 113) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
M3K12_MOUSE
|
||||||
| NC score | 0.000772 (rank : 114) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
ZF276_HUMAN
|
||||||
| NC score | 0.000027 (rank : 115) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
ZN206_HUMAN
|
||||||
| NC score | -0.000967 (rank : 116) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96SZ4 | Gene names | ZNF206, ZSCAN10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 206 (Zinc finger and SCAN domain-containing protein 10). | |||||
|
ZNF18_MOUSE
|
||||||
| NC score | -0.001512 (rank : 117) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810A1, Q9D972 | Gene names | Znf18, Zfp535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 18 (Zinc finger protein 535). | |||||