Please be patient as the page loads
|
TBX2_HUMAN
|
||||||
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TBX2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 195 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TBX2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990661 (rank : 2) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q60707 | Gene names | Tbx2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TBX3_MOUSE
|
||||||
| θ value | 5.3931e-154 (rank : 3) | NC score | 0.984151 (rank : 3) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70324, Q60708, Q8BWH7 | Gene names | Tbx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
TBX3_HUMAN
|
||||||
| θ value | 1.91815e-151 (rank : 4) | NC score | 0.978805 (rank : 4) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
TBX4_MOUSE
|
||||||
| θ value | 3.13423e-69 (rank : 5) | NC score | 0.956475 (rank : 12) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70325, Q8R5F6 | Gene names | Tbx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
TBX4_HUMAN
|
||||||
| θ value | 1.90147e-66 (rank : 6) | NC score | 0.956078 (rank : 14) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P57082 | Gene names | TBX4 | |||
|
Domain Architecture |
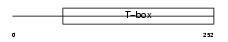
|
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
TBX5_HUMAN
|
||||||
| θ value | 2.74572e-65 (rank : 7) | NC score | 0.956458 (rank : 13) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99593, O15301, Q9Y4I2 | Gene names | TBX5 | |||
|
Domain Architecture |
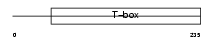
|
|||||
| Description | T-box transcription factor TBX5 (T-box protein 5). | |||||
|
TBX5_MOUSE
|
||||||
| θ value | 2.74572e-65 (rank : 8) | NC score | 0.956015 (rank : 15) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P70326, Q9R003 | Gene names | Tbx5 | |||
|
Domain Architecture |
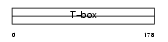
|
|||||
| Description | T-box transcription factor TBX5 (T-box protein 5). | |||||
|
TBX20_MOUSE
|
||||||
| θ value | 3.58603e-65 (rank : 9) | NC score | 0.970682 (rank : 5) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ES03, Q9ESX1 | Gene names | Tbx20, Tbx12 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX20 (T-box protein 20). | |||||
|
TBX6_HUMAN
|
||||||
| θ value | 5.7252e-63 (rank : 10) | NC score | 0.962834 (rank : 8) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95947, Q8TAS4 | Gene names | TBX6 | |||
|
Domain Architecture |
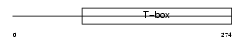
|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
TBX6_MOUSE
|
||||||
| θ value | 3.71096e-62 (rank : 11) | NC score | 0.959638 (rank : 11) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70327 | Gene names | Tbx6 | |||
|
Domain Architecture |
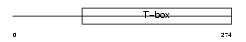
|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
TBX1_MOUSE
|
||||||
| θ value | 1.84173e-61 (rank : 12) | NC score | 0.964521 (rank : 7) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70323, Q60706, Q99MP0, Q99P22 | Gene names | Tbx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX1 (T-box protein 1) (Testis-specific T- box protein). | |||||
|
TBX1_HUMAN
|
||||||
| θ value | 4.10295e-61 (rank : 13) | NC score | 0.964655 (rank : 6) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O43435, O43436, Q96RJ2 | Gene names | TBX1 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX1 (T-box protein 1) (Testis-specific T- box protein). | |||||
|
TBX10_MOUSE
|
||||||
| θ value | 9.1404e-61 (rank : 14) | NC score | 0.961315 (rank : 10) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q810F8, O54841, Q6QH75 | Gene names | Tbx10, Tbx13, Tbx7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10) (T-box protein 13) (MmTBX7). | |||||
|
TBX10_HUMAN
|
||||||
| θ value | 2.03627e-60 (rank : 15) | NC score | 0.961767 (rank : 9) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75333, Q86XS3 | Gene names | TBX10, TBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10). | |||||
|
TBX18_HUMAN
|
||||||
| θ value | 2.65945e-60 (rank : 16) | NC score | 0.944793 (rank : 19) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |
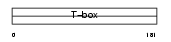
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
TBX18_MOUSE
|
||||||
| θ value | 8.55514e-59 (rank : 17) | NC score | 0.948146 (rank : 18) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EPZ6 | Gene names | Tbx18 | |||
|
Domain Architecture |
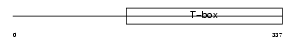
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
TBX15_HUMAN
|
||||||
| θ value | 6.77864e-56 (rank : 18) | NC score | 0.944387 (rank : 20) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SF7, Q5T9S7 | Gene names | TBX15, TBX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15). | |||||
|
TBX15_MOUSE
|
||||||
| θ value | 2.57589e-55 (rank : 19) | NC score | 0.944337 (rank : 21) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |
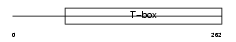
|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
TBX22_MOUSE
|
||||||
| θ value | 6.34462e-54 (rank : 20) | NC score | 0.951585 (rank : 17) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K402 | Gene names | Tbx22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX22 (T-box protein 22). | |||||
|
TBX22_HUMAN
|
||||||
| θ value | 3.1488e-53 (rank : 21) | NC score | 0.952017 (rank : 16) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y458, Q96LC0, Q9HBF1 | Gene names | TBX22, TBOX22 | |||
|
Domain Architecture |
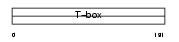
|
|||||
| Description | T-box transcription factor TBX22 (T-box protein 22). | |||||
|
BRAC_HUMAN
|
||||||
| θ value | 8.57503e-51 (rank : 22) | NC score | 0.939998 (rank : 23) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15178 | Gene names | T | |||
|
Domain Architecture |
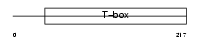
|
|||||
| Description | Brachyury protein (T protein). | |||||
|
BRAC_MOUSE
|
||||||
| θ value | 2.49495e-50 (rank : 23) | NC score | 0.941071 (rank : 22) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P20293 | Gene names | T | |||
|
Domain Architecture |

|
|||||
| Description | Brachyury protein (T protein). | |||||
|
TBX21_HUMAN
|
||||||
| θ value | 2.58187e-47 (rank : 24) | NC score | 0.929282 (rank : 27) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
TBR1_HUMAN
|
||||||
| θ value | 1.28137e-46 (rank : 25) | NC score | 0.925543 (rank : 29) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16650, Q53TH0 | Gene names | TBR1 | |||
|
Domain Architecture |
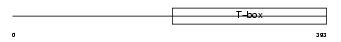
|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
TBR1_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 26) | NC score | 0.925498 (rank : 30) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q64336 | Gene names | Tbr1 | |||
|
Domain Architecture |
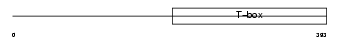
|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
TBX21_MOUSE
|
||||||
| θ value | 2.85459e-46 (rank : 27) | NC score | 0.932377 (rank : 26) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JKD8, Q9R0A6 | Gene names | Tbx21, Tbet, Tblym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
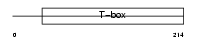
EOMES_HUMAN
|
||||||
| θ value | 6.35935e-46 (rank : 28) | NC score | 0.926699 (rank : 28) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |
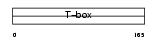
|
|||||
| Description | Eomesodermin homolog. | |||||
|
EOMES_MOUSE
|
||||||
| θ value | 4.122e-45 (rank : 29) | NC score | 0.925042 (rank : 31) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54839, Q9QYG7 | Gene names | Eomes, Tbr2 | |||
|
Domain Architecture |
|
|||||
| Description | Eomesodermin homolog. | |||||
|
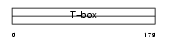
TBX19_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 30) | NC score | 0.937854 (rank : 24) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60806 | Gene names | TBX19, TPIT | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
TBX19_MOUSE
|
||||||
| θ value | 9.18288e-45 (rank : 31) | NC score | 0.934553 (rank : 25) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99ME7 | Gene names | Tbx19, Tpit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 32) | NC score | 0.039164 (rank : 37) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 33) | NC score | 0.037992 (rank : 38) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
EGLN1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 34) | NC score | 0.026879 (rank : 47) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
FOXC1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 35) | NC score | 0.012962 (rank : 105) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |
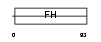
|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 36) | NC score | 0.040855 (rank : 34) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
PHF22_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 37) | NC score | 0.027826 (rank : 44) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
SN1L2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 38) | NC score | -0.000353 (rank : 183) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
MYL3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.012146 (rank : 112) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
SC5A3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.025721 (rank : 51) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
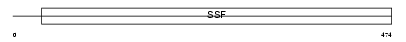
| SwissProt Accessions | Q9JKZ2 | Gene names | Slc5a3 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/myo-inositol cotransporter (Na(+)/myo-inositol cotransporter). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 41) | NC score | 0.011160 (rank : 121) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.008021 (rank : 145) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 43) | NC score | 0.022069 (rank : 63) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 44) | NC score | 0.044806 (rank : 32) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 0.163984 (rank : 45) | NC score | 0.040370 (rank : 36) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
INSM1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 46) | NC score | 0.008940 (rank : 139) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 683 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01101 | Gene names | INSM1, IA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
DPOD3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.025123 (rank : 56) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.018634 (rank : 75) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
JIP2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.017445 (rank : 80) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
TM16H_MOUSE
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.021120 (rank : 65) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PB70, Q69ZE4 | Gene names | Tmem16h, Kiaa1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.014605 (rank : 97) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.019289 (rank : 70) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.017227 (rank : 82) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
K0586_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.026273 (rank : 48) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
SRBS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.015958 (rank : 91) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.010864 (rank : 123) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
ASTL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.015487 (rank : 95) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.024414 (rank : 57) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
CO027_HUMAN
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.027784 (rank : 45) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2M3C6, Q8N993, Q96LL5 | Gene names | C15orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf27. | |||||
|
GNAQ_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.010394 (rank : 131) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
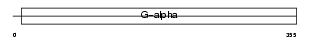
| SwissProt Accessions | P50148, O15108, Q13462, Q92471, Q9BZB9 | Gene names | GNAQ, GAQ | |||
|
Domain Architecture |
|
|||||
| Description | Guanine nucleotide-binding protein G(q) subunit alpha (Guanine nucleotide-binding protein alpha-q). | |||||
|
GNAQ_MOUSE
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.010384 (rank : 132) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
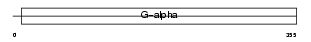
| SwissProt Accessions | P21279, Q6PFF5 | Gene names | Gnaq | |||
|
Domain Architecture |
|
|||||
| Description | Guanine nucleotide-binding protein G(q) subunit alpha (Guanine nucleotide-binding protein alpha-q). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.008845 (rank : 140) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.023482 (rank : 60) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TLX2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.009477 (rank : 136) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43763, Q9UQ48 | Gene names | TLX2, HOX11L1, NCX | |||
|
Domain Architecture |

|
|||||
| Description | T-cell leukemia homeobox protein 2 (Homeobox protein Hox-11L1) (Neural crest homeobox protein). | |||||
|
HXB4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.003721 (rank : 164) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10284, Q3UZB4, Q4VBG0 | Gene names | Hoxb4, Hox-2.6, Hoxb-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2.6). | |||||
|
IF39_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.025465 (rank : 53) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | -0.000367 (rank : 184) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
SNUT3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.035088 (rank : 39) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
|
FOXC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.010504 (rank : 129) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
FOXP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.011632 (rank : 114) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
GP124_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.003700 (rank : 165) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PE1, Q8N3R1, Q8TEM3, Q96KB2, Q9P1Z7, Q9UFY4 | Gene names | GPR124, KIAA1531, TEM5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
IKBL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.019278 (rank : 71) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
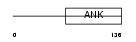
| SwissProt Accessions | O88995, Q91X16, Q9R1Y3 | Gene names | Nfkbil1, Ikbl | |||
|
Domain Architecture |
|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
LIPA3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.008408 (rank : 142) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
LYOX_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.010240 (rank : 133) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.018302 (rank : 77) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.026027 (rank : 50) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TEX28_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.016070 (rank : 88) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15482 | Gene names | CXorf2, TEX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific protein TEX28. | |||||
|
BAT3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.018519 (rank : 76) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
ERBB3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | -0.000472 (rank : 185) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
HIC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | -0.002301 (rank : 192) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
LRFN3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.002088 (rank : 175) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLY3 | Gene names | Lrfn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 3 precursor. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.004714 (rank : 160) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.034039 (rank : 40) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SC5A3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.018945 (rank : 74) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53794, O43489 | Gene names | SLC5A3 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/myo-inositol cotransporter (Na(+)/myo-inositol cotransporter). | |||||
|
STK6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | -0.002582 (rank : 193) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97477, O35624, Q91YU4 | Gene names | Stk6, Ark1, Aurka, Ayk1, Iak1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Aurora-family kinase 1) (Aurora/IPL1-related kinase 1) (Ipl1- and aurora-related kinase 1) (Aurora-A) (Serine/threonine-protein kinase Ayk1). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | -0.001725 (rank : 191) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.010713 (rank : 125) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.027207 (rank : 46) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.010721 (rank : 124) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.011045 (rank : 122) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
HASP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.013360 (rank : 102) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TF76, Q5U5K3, Q96MN1, Q9BXS7 | Gene names | GSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Haspin (EC 2.7.11.1) (Haploid germ cell-specific nuclear protein kinase) (H-Haspin) (Germ cell-specific gene 2 protein). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.011274 (rank : 119) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.018194 (rank : 79) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.020544 (rank : 68) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
INVS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.008227 (rank : 143) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.005425 (rank : 157) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
NKX12_MOUSE
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.002891 (rank : 169) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P42580, Q9JHF2, Q9JKW5, Q9JKW6, Q9JKW9, Q9JKX0, Q9JKX1 | Gene names | Nkx1-2, Nkx1.1, Sax1 | |||
|
Domain Architecture |

|
|||||
| Description | NK1 transcription factor-related protein 2 (Homeobox protein SAX-1) (NKX-1.1). | |||||
|
PALB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.012515 (rank : 110) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86YC2, Q8N7Y6, Q8ND31, Q9H6W1 | Gene names | PALB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.040662 (rank : 35) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
UST_MOUSE
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.020649 (rank : 66) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUB6 | Gene names | Ust | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uronyl 2-sulfotransferase (EC 2.8.2.-). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.020591 (rank : 67) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
CI078_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.019047 (rank : 72) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ63, Q8WVU6, Q9NT39 | Gene names | C9orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 (Hepatocellular carcinoma-associated antigen 59). | |||||
|
CI078_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.018972 (rank : 73) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TQI7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 homolog. | |||||
|
CT128_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.013614 (rank : 101) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.001324 (rank : 178) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.015579 (rank : 93) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
HSF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.010427 (rank : 130) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
HXA10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.002770 (rank : 171) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P31260, O43370, O43605, Q15949 | Gene names | HOXA10, HOX1H | |||
|
Domain Architecture |
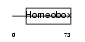
|
|||||
| Description | Homeobox protein Hox-A10 (Hox-1H) (Hox-1.8) (PL). | |||||
|
ICAL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.015092 (rank : 96) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.025465 (rank : 52) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
MICA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.006495 (rank : 152) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.020167 (rank : 69) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.016560 (rank : 86) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SNUT3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.028440 (rank : 43) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K194, Q8BRA8, Q9CSV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3. | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.017120 (rank : 83) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | -0.001253 (rank : 188) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
ZN750_HUMAN
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.012887 (rank : 107) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.030158 (rank : 42) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
BORG5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.010521 (rank : 127) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |
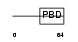
|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.004477 (rank : 162) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.012894 (rank : 106) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.011734 (rank : 113) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.010513 (rank : 128) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.008631 (rank : 141) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.026132 (rank : 49) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.034029 (rank : 41) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
NNP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.013148 (rank : 104) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56183, O35712, Q9ERE1, Q9JI07, Q9JK67, Q9JKU2 | Gene names | Nnp1 | |||
|
Domain Architecture |
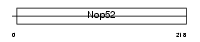
|
|||||
| Description | NNP-1 protein (Novel nuclear protein 1) (Nop52). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.004110 (rank : 163) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.003582 (rank : 167) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.009379 (rank : 138) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.015591 (rank : 92) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.025383 (rank : 54) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | 0.024247 (rank : 58) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
BEGIN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.004797 (rank : 159) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q68EF6 | Gene names | Begain | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.002985 (rank : 168) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
HXB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.002168 (rank : 174) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P0C1T1 | Gene names | Hoxb2, Hox-2.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein Hox-B2 (Hox-2.8). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.001262 (rank : 179) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
MECP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.016747 (rank : 85) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |
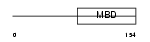
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.009957 (rank : 134) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | -0.001260 (rank : 189) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | -0.000656 (rank : 187) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.012404 (rank : 111) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.008139 (rank : 144) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
UST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.014156 (rank : 100) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2C2 | Gene names | UST, DS2ST | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uronyl 2-sulfotransferase (EC 2.8.2.-). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | 0.002459 (rank : 173) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.011601 (rank : 115) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.016023 (rank : 89) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.002883 (rank : 170) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
CDT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.012868 (rank : 108) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4E9, Q8BUQ1, Q8BX29, Q8R3V0, Q8R4E8 | Gene names | Cdt1, Ris2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP) (Retroviral insertion site 2 protein). | |||||
|
CR034_MOUSE
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.001029 (rank : 181) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.005634 (rank : 155) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.000985 (rank : 182) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.017412 (rank : 81) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.009807 (rank : 135) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
GTSE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.025250 (rank : 55) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
HIC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | -0.002985 (rank : 194) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14526 | Gene names | HIC1, ZBTB29 | |||
|
Domain Architecture |
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1) (Zinc finger and BTB domain-containing protein 29). | |||||
|
INSM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.005064 (rank : 158) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q63ZV0, Q9Z113 | Gene names | Insm1, Ia1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.007903 (rank : 148) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.004541 (rank : 161) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KLF13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | -0.001487 (rank : 190) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2Y9, Q9Y356 | Gene names | KLF13, BTEB3, NSLP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 13 (Transcription factor BTEB3) (Basic transcription element-binding protein 3) (BTE-binding protein 3) (RANTES factor of late activated T-lymphocytes 1) (RFLAT-1) (Transcription factor NSLP1) (Novel Sp1-like zinc finger transcription factor 1). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.007626 (rank : 149) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
LRFN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.001086 (rank : 180) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BTN0, Q6UY10 | Gene names | LRFN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 3 precursor. | |||||
|
NNP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | 0.011288 (rank : 118) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56182, Q9NST5 | Gene names | NNP1, NOP52 | |||
|
Domain Architecture |
|
|||||
| Description | NNP-1 protein (Novel nuclear protein 1) (Nucleolar protein Nop52) (D21S2056E). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.023708 (rank : 59) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 165) | NC score | 0.016010 (rank : 90) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RN169_HUMAN
|
||||||
| θ value | 6.88961 (rank : 166) | NC score | 0.006782 (rank : 151) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 167) | NC score | 0.001413 (rank : 176) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SFR16_MOUSE
|
||||||
| θ value | 6.88961 (rank : 168) | NC score | 0.014174 (rank : 99) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 169) | NC score | 0.011297 (rank : 117) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 170) | NC score | 0.018238 (rank : 78) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 6.88961 (rank : 171) | NC score | 0.011205 (rank : 120) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
WFS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 172) | NC score | 0.008020 (rank : 146) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56695, Q9Z276 | Gene names | Wfs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wolframin. | |||||
|
ZBTB4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 173) | NC score | -0.000623 (rank : 186) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.016158 (rank : 87) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.007581 (rank : 150) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.010529 (rank : 126) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.043223 (rank : 33) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.023288 (rank : 61) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.011435 (rank : 116) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.005929 (rank : 154) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
DUS9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.001373 (rank : 177) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 182) | NC score | 0.015537 (rank : 94) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 183) | NC score | 0.009461 (rank : 137) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PANK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 184) | NC score | 0.003670 (rank : 166) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZ23, Q5T7I2, Q8N7Q4, Q8TCR5, Q9BYW5, Q9HAF2 | Gene names | PANK2, C20orf48 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 185) | NC score | 0.014347 (rank : 98) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 186) | NC score | 0.006365 (rank : 153) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 187) | NC score | 0.013341 (rank : 103) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 188) | NC score | 0.016874 (rank : 84) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 189) | NC score | 0.022366 (rank : 62) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SYNJ2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 190) | NC score | 0.005471 (rank : 156) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15056, Q5TA13, Q5TA16, Q5TA19, Q86XK0, Q8IZA8, Q9H226 | Gene names | SYNJ2, KIAA0348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 191) | NC score | 0.012722 (rank : 109) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TERF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 192) | NC score | 0.007981 (rank : 147) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 193) | NC score | 0.021236 (rank : 64) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 8.99809 (rank : 194) | NC score | 0.002547 (rank : 172) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
ZN575_HUMAN
|
||||||
| θ value | 8.99809 (rank : 195) | NC score | -0.003974 (rank : 195) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||
|
TBX2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 195 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TBX2_MOUSE
|
||||||
| NC score | 0.990661 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q60707 | Gene names | Tbx2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TBX3_MOUSE
|
||||||
| NC score | 0.984151 (rank : 3) | θ value | 5.3931e-154 (rank : 3) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70324, Q60708, Q8BWH7 | Gene names | Tbx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
TBX3_HUMAN
|
||||||
| NC score | 0.978805 (rank : 4) | θ value | 1.91815e-151 (rank : 4) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
TBX20_MOUSE
|
||||||
| NC score | 0.970682 (rank : 5) | θ value | 3.58603e-65 (rank : 9) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ES03, Q9ESX1 | Gene names | Tbx20, Tbx12 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX20 (T-box protein 20). | |||||
|
TBX1_HUMAN
|
||||||
| NC score | 0.964655 (rank : 6) | θ value | 4.10295e-61 (rank : 13) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O43435, O43436, Q96RJ2 | Gene names | TBX1 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX1 (T-box protein 1) (Testis-specific T- box protein). | |||||
|
TBX1_MOUSE
|
||||||
| NC score | 0.964521 (rank : 7) | θ value | 1.84173e-61 (rank : 12) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70323, Q60706, Q99MP0, Q99P22 | Gene names | Tbx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX1 (T-box protein 1) (Testis-specific T- box protein). | |||||
|
TBX6_HUMAN
|
||||||
| NC score | 0.962834 (rank : 8) | θ value | 5.7252e-63 (rank : 10) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95947, Q8TAS4 | Gene names | TBX6 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
TBX10_HUMAN
|
||||||
| NC score | 0.961767 (rank : 9) | θ value | 2.03627e-60 (rank : 15) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75333, Q86XS3 | Gene names | TBX10, TBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10). | |||||
|
TBX10_MOUSE
|
||||||
| NC score | 0.961315 (rank : 10) | θ value | 9.1404e-61 (rank : 14) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q810F8, O54841, Q6QH75 | Gene names | Tbx10, Tbx13, Tbx7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10) (T-box protein 13) (MmTBX7). | |||||
|
TBX6_MOUSE
|
||||||
| NC score | 0.959638 (rank : 11) | θ value | 3.71096e-62 (rank : 11) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70327 | Gene names | Tbx6 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
TBX4_MOUSE
|
||||||
| NC score | 0.956475 (rank : 12) | θ value | 3.13423e-69 (rank : 5) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70325, Q8R5F6 | Gene names | Tbx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
TBX5_HUMAN
|
||||||
| NC score | 0.956458 (rank : 13) | θ value | 2.74572e-65 (rank : 7) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99593, O15301, Q9Y4I2 | Gene names | TBX5 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX5 (T-box protein 5). | |||||
|
TBX4_HUMAN
|
||||||
| NC score | 0.956078 (rank : 14) | θ value | 1.90147e-66 (rank : 6) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P57082 | Gene names | TBX4 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
TBX5_MOUSE
|
||||||
| NC score | 0.956015 (rank : 15) | θ value | 2.74572e-65 (rank : 8) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P70326, Q9R003 | Gene names | Tbx5 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX5 (T-box protein 5). | |||||
|
TBX22_HUMAN
|
||||||
| NC score | 0.952017 (rank : 16) | θ value | 3.1488e-53 (rank : 21) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y458, Q96LC0, Q9HBF1 | Gene names | TBX22, TBOX22 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX22 (T-box protein 22). | |||||
|
TBX22_MOUSE
|
||||||
| NC score | 0.951585 (rank : 17) | θ value | 6.34462e-54 (rank : 20) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K402 | Gene names | Tbx22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX22 (T-box protein 22). | |||||
|
TBX18_MOUSE
|
||||||
| NC score | 0.948146 (rank : 18) | θ value | 8.55514e-59 (rank : 17) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EPZ6 | Gene names | Tbx18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
TBX18_HUMAN
|
||||||
| NC score | 0.944793 (rank : 19) | θ value | 2.65945e-60 (rank : 16) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
TBX15_HUMAN
|
||||||
| NC score | 0.944387 (rank : 20) | θ value | 6.77864e-56 (rank : 18) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SF7, Q5T9S7 | Gene names | TBX15, TBX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15). | |||||
|
TBX15_MOUSE
|
||||||
| NC score | 0.944337 (rank : 21) | θ value | 2.57589e-55 (rank : 19) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
BRAC_MOUSE
|
||||||
| NC score | 0.941071 (rank : 22) | θ value | 2.49495e-50 (rank : 23) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P20293 | Gene names | T | |||
|
Domain Architecture |

|
|||||
| Description | Brachyury protein (T protein). | |||||
|
BRAC_HUMAN
|
||||||
| NC score | 0.939998 (rank : 23) | θ value | 8.57503e-51 (rank : 22) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15178 | Gene names | T | |||
|
Domain Architecture |

|
|||||
| Description | Brachyury protein (T protein). | |||||
|
TBX19_HUMAN
|
||||||
| NC score | 0.937854 (rank : 24) | θ value | 9.18288e-45 (rank : 30) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60806 | Gene names | TBX19, TPIT | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
TBX19_MOUSE
|
||||||
| NC score | 0.934553 (rank : 25) | θ value | 9.18288e-45 (rank : 31) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99ME7 | Gene names | Tbx19, Tpit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
TBX21_MOUSE
|
||||||
| NC score | 0.932377 (rank : 26) | θ value | 2.85459e-46 (rank : 27) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JKD8, Q9R0A6 | Gene names | Tbx21, Tbet, Tblym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
TBX21_HUMAN
|
||||||
| NC score | 0.929282 (rank : 27) | θ value | 2.58187e-47 (rank : 24) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
EOMES_HUMAN
|
||||||
| NC score | 0.926699 (rank : 28) | θ value | 6.35935e-46 (rank : 28) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |

|
|||||
| Description | Eomesodermin homolog. | |||||
|
TBR1_HUMAN
|
||||||
| NC score | 0.925543 (rank : 29) | θ value | 1.28137e-46 (rank : 25) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16650, Q53TH0 | Gene names | TBR1 | |||
|
Domain Architecture |

|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
TBR1_MOUSE
|
||||||
| NC score | 0.925498 (rank : 30) | θ value | 1.28137e-46 (rank : 26) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q64336 | Gene names | Tbr1 | |||
|
Domain Architecture |

|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
EOMES_MOUSE
|
||||||
| NC score | 0.925042 (rank : 31) | θ value | 4.122e-45 (rank : 29) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54839, Q9QYG7 | Gene names | Eomes, Tbr2 | |||
|
Domain Architecture |

|
|||||
| Description | Eomesodermin homolog. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.044806 (rank : 32) | θ value | 0.163984 (rank : 44) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.043223 (rank : 33) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.040855 (rank : 34) | θ value | 0.0736092 (rank : 36) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.040662 (rank : 35) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.040370 (rank : 36) | θ value | 0.163984 (rank : 45) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.039164 (rank : 37) | θ value | 0.000158464 (rank : 32) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.037992 (rank : 38) | θ value | 0.0330416 (rank : 33) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
SNUT3_HUMAN
|
||||||
| NC score | 0.035088 (rank : 39) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.034039 (rank : 40) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.034029 (rank : 41) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.030158 (rank : 42) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
SNUT3_MOUSE
|
||||||
| NC score | 0.028440 (rank : 43) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K194, Q8BRA8, Q9CSV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3. | |||||
|
PHF22_MOUSE
|
||||||
| NC score | 0.027826 (rank : 44) | θ value | 0.0961366 (rank : 37) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
CO027_HUMAN
|
||||||
| NC score | 0.027784 (rank : 45) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2M3C6, Q8N993, Q96LL5 | Gene names | C15orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf27. | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.027207 (rank : 46) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
EGLN1_MOUSE
|
||||||
| NC score | 0.026879 (rank : 47) | θ value | 0.0330416 (rank : 34) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
K0586_HUMAN
|
||||||
| NC score | 0.026273 (rank : 48) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.026132 (rank : 49) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.026027 (rank : 50) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SC5A3_MOUSE
|
||||||
| NC score | 0.025721 (rank : 51) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JKZ2 | Gene names | Slc5a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/myo-inositol cotransporter (Na(+)/myo-inositol cotransporter). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.025465 (rank : 52) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.025465 (rank : 53) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.025383 (rank : 54) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
GTSE1_HUMAN
|
||||||
| NC score | 0.025250 (rank : 55) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
DPOD3_HUMAN
|
||||||
| NC score | 0.025123 (rank : 56) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.024414 (rank : 57) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.024247 (rank : 58) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.023708 (rank : 59) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.023482 (rank : 60) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.023288 (rank : 61) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.022366 (rank : 62) | θ value | 8.99809 (rank : 189) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.022069 (rank : 63) | θ value | 0.163984 (rank : 43) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.021236 (rank : 64) | θ value | 8.99809 (rank : 193) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
TM16H_MOUSE
|
||||||
| NC score | 0.021120 (rank : 65) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PB70, Q69ZE4 | Gene names | Tmem16h, Kiaa1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
UST_MOUSE
|
||||||
| NC score | 0.020649 (rank : 66) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUB6 | Gene names | Ust | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uronyl 2-sulfotransferase (EC 2.8.2.-). | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.020591 (rank : 67) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.020544 (rank : 68) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.020167 (rank : 69) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.019289 (rank : 70) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
IKBL_MOUSE
|
||||||
| NC score | 0.019278 (rank : 71) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88995, Q91X16, Q9R1Y3 | Gene names | Nfkbil1, Ikbl | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
CI078_HUMAN
|
||||||
| NC score | 0.019047 (rank : 72) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ63, Q8WVU6, Q9NT39 | Gene names | C9orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 (Hepatocellular carcinoma-associated antigen 59). | |||||
|
CI078_MOUSE
|
||||||
| NC score | 0.018972 (rank : 73) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TQI7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 homolog. | |||||
|
SC5A3_HUMAN
|
||||||
| NC score | 0.018945 (rank : 74) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53794, O43489 | Gene names | SLC5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/myo-inositol cotransporter (Na(+)/myo-inositol cotransporter). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.018634 (rank : 75) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.018519 (rank : 76) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.018302 (rank : 77) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.018238 (rank : 78) | θ value | 6.88961 (rank : 170) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.018194 (rank : 79) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
JIP2_MOUSE
|
||||||
| NC score | 0.017445 (rank : 80) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.017412 (rank : 81) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.017227 (rank : 82) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.017120 (rank : 83) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.016874 (rank : 84) | θ value | 8.99809 (rank : 188) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MECP2_HUMAN
|
||||||
| NC score | 0.016747 (rank : 85) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.016560 (rank : 86) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.016158 (rank : 87) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
TEX28_HUMAN
|
||||||
| NC score | 0.016070 (rank : 88) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15482 | Gene names | CXorf2, TEX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific protein TEX28. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.016023 (rank : 89) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.016010 (rank : 90) | θ value | 6.88961 (rank : 165) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SRBS1_HUMAN
|
||||||
| NC score | 0.015958 (rank : 91) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.015591 (rank : 92) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.015579 (rank : 93) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.015537 (rank : 94) | θ value | 8.99809 (rank : 182) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
ASTL_HUMAN
|
||||||
| NC score | 0.015487 (rank : 95) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
ICAL_MOUSE
|
||||||
| NC score | 0.015092 (rank : 96) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.014605 (rank : 97) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.014347 (rank : 98) | θ value | 8.99809 (rank : 185) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SFR16_MOUSE
|
||||||
| NC score | 0.014174 (rank : 99) | θ value | 6.88961 (rank : 168) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
UST_HUMAN
|
||||||
| NC score | 0.014156 (rank : 100) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2C2 | Gene names | UST, DS2ST | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uronyl 2-sulfotransferase (EC 2.8.2.-). | |||||
|
CT128_HUMAN
|
||||||
| NC score | 0.013614 (rank : 101) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
HASP_HUMAN
|
||||||
| NC score | 0.013360 (rank : 102) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TF76, Q5U5K3, Q96MN1, Q9BXS7 | Gene names | GSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Haspin (EC 2.7.11.1) (Haploid germ cell-specific nuclear protein kinase) (H-Haspin) (Germ cell-specific gene 2 protein). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.013341 (rank : 103) | θ value | 8.99809 (rank : 187) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NNP1_MOUSE
|
||||||
| NC score | 0.013148 (rank : 104) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56183, O35712, Q9ERE1, Q9JI07, Q9JK67, Q9JKU2 | Gene names | Nnp1 | |||
|
Domain Architecture |

|
|||||
| Description | NNP-1 protein (Novel nuclear protein 1) (Nop52). | |||||
|
FOXC1_HUMAN
|
||||||
| NC score | 0.012962 (rank : 105) | θ value | 0.0736092 (rank : 35) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.012894 (rank : 106) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
ZN750_HUMAN
|
||||||
| NC score | 0.012887 (rank : 107) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CDT1_MOUSE
|
||||||
| NC score | 0.012868 (rank : 108) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4E9, Q8BUQ1, Q8BX29, Q8R3V0, Q8R4E8 | Gene names | Cdt1, Ris2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP) (Retroviral insertion site 2 protein). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.012722 (rank : 109) | θ value | 8.99809 (rank : 191) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PALB2_HUMAN
|
||||||
| NC score | 0.012515 (rank : 110) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86YC2, Q8N7Y6, Q8ND31, Q9H6W1 | Gene names | PALB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.012404 (rank : 111) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
MYL3_MOUSE
|
||||||
| NC score | 0.012146 (rank : 112) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.011734 (rank : 113) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
FOXP4_HUMAN
|
||||||
| NC score | 0.011632 (rank : 114) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.011601 (rank : 115) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.011435 (rank : 116) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.011297 (rank : 117) | θ value | 6.88961 (rank : 169) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
NNP1_HUMAN
|
||||||
| NC score | 0.011288 (rank : 118) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56182, Q9NST5 | Gene names | NNP1, NOP52 | |||
|
Domain Architecture |

|
|||||
| Description | NNP-1 protein (Novel nuclear protein 1) (Nucleolar protein Nop52) (D21S2056E). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.011274 (rank : 119) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.011205 (rank : 120) | θ value | 6.88961 (rank : 171) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.011160 (rank : 121) | θ value | 0.125558 (rank : 41) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.011045 (rank : 122) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.010864 (rank : 123) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.010721 (rank : 124) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.010713 (rank : 125) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.010529 (rank : 126) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
BORG5_HUMAN
|
||||||
| NC score | 0.010521 (rank : 127) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.010513 (rank : 128) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
FOXC1_MOUSE
|
||||||
| NC score | 0.010504 (rank : 129) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
HSF4_HUMAN
|
||||||
| NC score | 0.010427 (rank : 130) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
GNAQ_HUMAN
|
||||||
| NC score | 0.010394 (rank : 131) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50148, O15108, Q13462, Q92471, Q9BZB9 | Gene names | GNAQ, GAQ | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(q) subunit alpha (Guanine nucleotide-binding protein alpha-q). | |||||
|
GNAQ_MOUSE
|
||||||
| NC score | 0.010384 (rank : 132) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21279, Q6PFF5 | Gene names | Gnaq | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(q) subunit alpha (Guanine nucleotide-binding protein alpha-q). | |||||
|
LYOX_MOUSE
|
||||||
| NC score | 0.010240 (rank : 133) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.009957 (rank : 134) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.009807 (rank : 135) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
TLX2_HUMAN
|
||||||
| NC score | 0.009477 (rank : 136) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43763, Q9UQ48 | Gene names | TLX2, HOX11L1, NCX | |||
|
Domain Architecture |

|
|||||
| Description | T-cell leukemia homeobox protein 2 (Homeobox protein Hox-11L1) (Neural crest homeobox protein). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.009461 (rank : 137) | θ value | 8.99809 (rank : 183) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.009379 (rank : 138) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
INSM1_HUMAN
|
||||||
| NC score | 0.008940 (rank : 139) | θ value | 0.21417 (rank : 46) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 683 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01101 | Gene names | INSM1, IA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.008845 (rank : 140) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.008631 (rank : 141) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
LIPA3_MOUSE
|
||||||
| NC score | 0.008408 (rank : 142) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.008227 (rank : 143) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.008139 (rank : 144) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.008021 (rank : 145) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
WFS1_MOUSE
|
||||||
| NC score | 0.008020 (rank : 146) | θ value | 6.88961 (rank : 172) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56695, Q9Z276 | Gene names | Wfs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wolframin. | |||||
|
TERF2_MOUSE
|
||||||
| NC score | 0.007981 (rank : 147) | θ value | 8.99809 (rank : 192) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.007903 (rank : 148) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.007626 (rank : 149) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.007581 (rank : 150) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
RN169_HUMAN
|
||||||
| NC score | 0.006782 (rank : 151) | θ value | 6.88961 (rank : 166) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
MICA1_MOUSE
|
||||||
| NC score | 0.006495 (rank : 152) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.006365 (rank : 153) | θ value | 8.99809 (rank : 186) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.005929 (rank : 154) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.005634 (rank : 155) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
SYNJ2_HUMAN
|
||||||
| NC score | 0.005471 (rank : 156) | θ value | 8.99809 (rank : 190) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15056, Q5TA13, Q5TA16, Q5TA19, Q86XK0, Q8IZA8, Q9H226 | Gene names | SYNJ2, KIAA0348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.005425 (rank : 157) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
INSM1_MOUSE
|
||||||
| NC score | 0.005064 (rank : 158) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q63ZV0, Q9Z113 | Gene names | Insm1, Ia1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
BEGIN_MOUSE
|
||||||
| NC score | 0.004797 (rank : 159) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q68EF6 | Gene names | Begain | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.004714 (rank : 160) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.004541 (rank : 161) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.004477 (rank : 162) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.004110 (rank : 163) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
HXB4_MOUSE
|
||||||
| NC score | 0.003721 (rank : 164) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10284, Q3UZB4, Q4VBG0 | Gene names | Hoxb4, Hox-2.6, Hoxb-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2.6). | |||||
|
GP124_HUMAN
|
||||||
| NC score | 0.003700 (rank : 165) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PE1, Q8N3R1, Q8TEM3, Q96KB2, Q9P1Z7, Q9UFY4 | Gene names | GPR124, KIAA1531, TEM5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
PANK2_HUMAN
|
||||||
| NC score | 0.003670 (rank : 166) | θ value | 8.99809 (rank : 184) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZ23, Q5T7I2, Q8N7Q4, Q8TCR5, Q9BYW5, Q9HAF2 | Gene names | PANK2, C20orf48 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.003582 (rank : 167) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.002985 (rank : 168) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
NKX12_MOUSE
|
||||||
| NC score | 0.002891 (rank : 169) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P42580, Q9JHF2, Q9JKW5, Q9JKW6, Q9JKW9, Q9JKX0, Q9JKX1 | Gene names | Nkx1-2, Nkx1.1, Sax1 | |||
|
Domain Architecture |

|
|||||
| Description | NK1 transcription factor-related protein 2 (Homeobox protein SAX-1) (NKX-1.1). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.002883 (rank : 170) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
HXA10_HUMAN
|
||||||
| NC score | 0.002770 (rank : 171) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P31260, O43370, O43605, Q15949 | Gene names | HOXA10, HOX1H | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A10 (Hox-1H) (Hox-1.8) (PL). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.002547 (rank : 172) | θ value | 8.99809 (rank : 194) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.002459 (rank : 173) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
HXB2_MOUSE
|
||||||
| NC score | 0.002168 (rank : 174) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P0C1T1 | Gene names | Hoxb2, Hox-2.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein Hox-B2 (Hox-2.8). | |||||
|
LRFN3_MOUSE
|
||||||
| NC score | 0.002088 (rank : 175) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLY3 | Gene names | Lrfn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 3 precursor. | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.001413 (rank : 176) | θ value | 6.88961 (rank : 167) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
DUS9_HUMAN
|
||||||
| NC score | 0.001373 (rank : 177) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.001324 (rank : 178) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.001262 (rank : 179) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
LRFN3_HUMAN
|
||||||
| NC score | 0.001086 (rank : 180) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BTN0, Q6UY10 | Gene names | LRFN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 3 precursor. | |||||
|
CR034_MOUSE
|
||||||
| NC score | 0.001029 (rank : 181) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.000985 (rank : 182) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
SN1L2_HUMAN
|
||||||
| NC score | -0.000353 (rank : 183) | θ value | 0.0961366 (rank : 38) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | -0.000367 (rank : 184) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
ERBB3_MOUSE
|
||||||
| NC score | -0.000472 (rank : 185) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
ZBTB4_HUMAN
|
||||||
| NC score | -0.000623 (rank : 186) | θ value | 6.88961 (rank : 173) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | -0.000656 (rank : 187) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
ULK1_HUMAN
|
||||||
| NC score | -0.001253 (rank : 188) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | -0.001260 (rank : 189) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
KLF13_HUMAN
|
||||||
| NC score | -0.001487 (rank : 190) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2Y9, Q9Y356 | Gene names | KLF13, BTEB3, NSLP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 13 (Transcription factor BTEB3) (Basic transcription element-binding protein 3) (BTE-binding protein 3) (RANTES factor of late activated T-lymphocytes 1) (RFLAT-1) (Transcription factor NSLP1) (Novel Sp1-like zinc finger transcription factor 1). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | -0.001725 (rank : 191) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
HIC1_MOUSE
|
||||||
| NC score | -0.002301 (rank : 192) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
STK6_MOUSE
|
||||||
| NC score | -0.002582 (rank : 193) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97477, O35624, Q91YU4 | Gene names | Stk6, Ark1, Aurka, Ayk1, Iak1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Aurora-family kinase 1) (Aurora/IPL1-related kinase 1) (Ipl1- and aurora-related kinase 1) (Aurora-A) (Serine/threonine-protein kinase Ayk1). | |||||
|
HIC1_HUMAN
|
||||||
| NC score | -0.002985 (rank : 194) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14526 | Gene names | HIC1, ZBTB29 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1) (Zinc finger and BTB domain-containing protein 29). | |||||
|
ZN575_HUMAN
|
||||||
| NC score | -0.003974 (rank : 195) | θ value | 8.99809 (rank : 195) | |||
| Query Neighborhood Hits | 195 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||