Please be patient as the page loads
|
TBX3_HUMAN
|
||||||
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TBX3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
TBX3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992510 (rank : 2) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70324, Q60708, Q8BWH7 | Gene names | Tbx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
TBX2_HUMAN
|
||||||
| θ value | 1.91815e-151 (rank : 3) | NC score | 0.978805 (rank : 4) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TBX2_MOUSE
|
||||||
| θ value | 2.19465e-147 (rank : 4) | NC score | 0.983041 (rank : 3) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60707 | Gene names | Tbx2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TBX4_MOUSE
|
||||||
| θ value | 6.76295e-64 (rank : 5) | NC score | 0.954109 (rank : 13) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P70325, Q8R5F6 | Gene names | Tbx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
TBX4_HUMAN
|
||||||
| θ value | 9.76566e-63 (rank : 6) | NC score | 0.953761 (rank : 14) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P57082 | Gene names | TBX4 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
TBX5_MOUSE
|
||||||
| θ value | 2.17557e-62 (rank : 7) | NC score | 0.953615 (rank : 15) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70326, Q9R003 | Gene names | Tbx5 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX5 (T-box protein 5). | |||||
|
TBX5_HUMAN
|
||||||
| θ value | 8.26713e-62 (rank : 8) | NC score | 0.954144 (rank : 12) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99593, O15301, Q9Y4I2 | Gene names | TBX5 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX5 (T-box protein 5). | |||||
|
TBX20_MOUSE
|
||||||
| θ value | 4.10295e-61 (rank : 9) | NC score | 0.968219 (rank : 5) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ES03, Q9ESX1 | Gene names | Tbx20, Tbx12 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX20 (T-box protein 20). | |||||
|
TBX1_MOUSE
|
||||||
| θ value | 1.61343e-57 (rank : 10) | NC score | 0.961830 (rank : 7) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70323, Q60706, Q99MP0, Q99P22 | Gene names | Tbx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX1 (T-box protein 1) (Testis-specific T- box protein). | |||||
|
TBX1_HUMAN
|
||||||
| θ value | 6.13105e-57 (rank : 11) | NC score | 0.962020 (rank : 6) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43435, O43436, Q96RJ2 | Gene names | TBX1 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX1 (T-box protein 1) (Testis-specific T- box protein). | |||||
|
TBX6_HUMAN
|
||||||
| θ value | 1.78386e-56 (rank : 12) | NC score | 0.959130 (rank : 9) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95947, Q8TAS4 | Gene names | TBX6 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
TBX10_HUMAN
|
||||||
| θ value | 2.32979e-56 (rank : 13) | NC score | 0.959197 (rank : 8) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75333, Q86XS3 | Gene names | TBX10, TBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10). | |||||
|
TBX10_MOUSE
|
||||||
| θ value | 6.77864e-56 (rank : 14) | NC score | 0.958444 (rank : 10) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q810F8, O54841, Q6QH75 | Gene names | Tbx10, Tbx13, Tbx7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10) (T-box protein 13) (MmTBX7). | |||||
|
TBX18_HUMAN
|
||||||
| θ value | 3.36421e-55 (rank : 15) | NC score | 0.941214 (rank : 19) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
TBX18_MOUSE
|
||||||
| θ value | 5.73848e-55 (rank : 16) | NC score | 0.944507 (rank : 18) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EPZ6 | Gene names | Tbx18 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
TBX6_MOUSE
|
||||||
| θ value | 9.78833e-55 (rank : 17) | NC score | 0.955743 (rank : 11) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70327 | Gene names | Tbx6 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
TBX15_HUMAN
|
||||||
| θ value | 2.66562e-52 (rank : 18) | NC score | 0.941107 (rank : 20) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SF7, Q5T9S7 | Gene names | TBX15, TBX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15). | |||||
|
TBX15_MOUSE
|
||||||
| θ value | 1.01294e-51 (rank : 19) | NC score | 0.940979 (rank : 21) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
TBX22_HUMAN
|
||||||
| θ value | 5.20228e-48 (rank : 20) | NC score | 0.948186 (rank : 16) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y458, Q96LC0, Q9HBF1 | Gene names | TBX22, TBOX22 | |||
|
Domain Architecture |
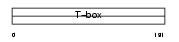
|
|||||
| Description | T-box transcription factor TBX22 (T-box protein 22). | |||||
|
TBX22_MOUSE
|
||||||
| θ value | 6.7944e-48 (rank : 21) | NC score | 0.947707 (rank : 17) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K402 | Gene names | Tbx22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX22 (T-box protein 22). | |||||
|
BRAC_HUMAN
|
||||||
| θ value | 7.51208e-47 (rank : 22) | NC score | 0.936549 (rank : 23) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15178 | Gene names | T | |||
|
Domain Architecture |
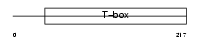
|
|||||
| Description | Brachyury protein (T protein). | |||||
|
BRAC_MOUSE
|
||||||
| θ value | 2.18568e-46 (rank : 23) | NC score | 0.937562 (rank : 22) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20293 | Gene names | T | |||
|
Domain Architecture |

|
|||||
| Description | Brachyury protein (T protein). | |||||
|
TBR1_HUMAN
|
||||||
| θ value | 1.85029e-45 (rank : 24) | NC score | 0.923934 (rank : 30) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16650, Q53TH0 | Gene names | TBR1 | |||
|
Domain Architecture |
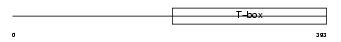
|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
TBR1_MOUSE
|
||||||
| θ value | 1.85029e-45 (rank : 25) | NC score | 0.923961 (rank : 29) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q64336 | Gene names | Tbr1 | |||
|
Domain Architecture |
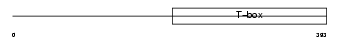
|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
TBX21_HUMAN
|
||||||
| θ value | 3.48949e-44 (rank : 26) | NC score | 0.926462 (rank : 27) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
TBX21_MOUSE
|
||||||
| θ value | 7.77379e-44 (rank : 27) | NC score | 0.929228 (rank : 26) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKD8, Q9R0A6 | Gene names | Tbx21, Tbet, Tblym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
EOMES_HUMAN
|
||||||
| θ value | 6.58091e-43 (rank : 28) | NC score | 0.924568 (rank : 28) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |
|
|||||
| Description | Eomesodermin homolog. | |||||
|
EOMES_MOUSE
|
||||||
| θ value | 6.58091e-43 (rank : 29) | NC score | 0.922929 (rank : 31) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54839, Q9QYG7 | Gene names | Eomes, Tbr2 | |||
|
Domain Architecture |
|
|||||
| Description | Eomesodermin homolog. | |||||
|
TBX19_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 30) | NC score | 0.933570 (rank : 24) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60806 | Gene names | TBX19, TPIT | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
TBX19_MOUSE
|
||||||
| θ value | 8.04462e-41 (rank : 31) | NC score | 0.930321 (rank : 25) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99ME7 | Gene names | Tbx19, Tpit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 32) | NC score | 0.022815 (rank : 50) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 33) | NC score | 0.040210 (rank : 36) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 34) | NC score | 0.053534 (rank : 32) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
BASP_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 35) | NC score | 0.052541 (rank : 33) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 36) | NC score | 0.042453 (rank : 35) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CIR_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 37) | NC score | 0.052059 (rank : 34) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 38) | NC score | 0.031756 (rank : 42) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
BASP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.038287 (rank : 37) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TACC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.015701 (rank : 68) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.025680 (rank : 45) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.017641 (rank : 66) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.010703 (rank : 86) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
TAOK2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | -0.001875 (rank : 118) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.013310 (rank : 73) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.023513 (rank : 48) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
TULP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.018861 (rank : 62) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.013356 (rank : 72) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.036573 (rank : 38) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.010570 (rank : 87) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.012684 (rank : 76) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.004901 (rank : 103) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
TLE4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.010194 (rank : 88) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.035228 (rank : 39) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.004673 (rank : 104) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.020526 (rank : 55) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.018540 (rank : 63) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.028287 (rank : 44) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.017983 (rank : 65) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.021578 (rank : 52) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TLE4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.009880 (rank : 89) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.015531 (rank : 69) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
K1196_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.009564 (rank : 91) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.023028 (rank : 49) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.011754 (rank : 78) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PO2F1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.007688 (rank : 97) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P25425, Q61994, Q63891, Q6Y681, Q7TSD0, Q8BT04, Q8K570, Q99JH0, Q9WTZ4, Q9WTZ5, Q9WTZ6 | Gene names | Pou2f1, Oct-1, Otf-1, Otf1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.020401 (rank : 56) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.020401 (rank : 57) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
SAFB2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.009867 (rank : 90) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.034075 (rank : 40) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DIP2C_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.011297 (rank : 79) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2E4 | Gene names | DIP2C, KIAA0934 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog C. | |||||
|
GCM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.016986 (rank : 67) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NP62, Q5T0X0, Q99468, Q9P1X3 | Gene names | GCM1, GCMA | |||
|
Domain Architecture |
|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (hGCMa). | |||||
|
HNRPF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.011244 (rank : 81) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52597, Q5T0N2, Q96AU2 | Gene names | HNRPF | |||
|
Domain Architecture |
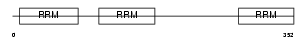
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein F (hnRNP F) (Nucleolin-like protein mcs94-1). | |||||
|
HNRPF_MOUSE
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.011274 (rank : 80) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2X1, Q5FWK2, Q8BVU8, Q8K2U9, Q8R0E7 | Gene names | Hnrpf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein F (hnRNP F). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.004256 (rank : 106) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.024813 (rank : 46) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.032993 (rank : 41) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.001215 (rank : 113) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BIN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.010873 (rank : 85) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.020192 (rank : 58) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.024109 (rank : 47) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
RNF12_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.008428 (rank : 93) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
SCG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.020020 (rank : 59) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.021242 (rank : 53) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.000242 (rank : 116) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.018269 (rank : 64) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.000833 (rank : 114) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
ADA2B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | -0.003751 (rank : 121) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 881 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18089, Q4TUH9, Q53RF2, Q9BZK0 | Gene names | ADRA2B | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2B adrenergic receptor (Alpha-2B adrenoceptor) (Alpha-2B adrenoreceptor) (Alpha-2 adrenergic receptor subtype C2). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.012208 (rank : 77) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.019091 (rank : 61) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NIPA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.011167 (rank : 82) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.010956 (rank : 83) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
S27A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.004017 (rank : 107) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PCB7 | Gene names | SLC27A1, ACSVL5, FATP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Long-chain fatty acid transport protein 1 (EC 6.2.1.-) (Fatty acid transport protein 1) (FATP-1) (Solute carrier family 27 member 1). | |||||
|
SH2D3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.005509 (rank : 102) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
TAF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.006018 (rank : 100) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.008387 (rank : 94) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.008347 (rank : 95) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.031420 (rank : 43) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.007045 (rank : 98) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.010946 (rank : 84) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.014435 (rank : 71) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
OAS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.005799 (rank : 101) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.013119 (rank : 74) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
PO4F3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.002831 (rank : 112) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63955 | Gene names | Pou4f3, Brn-3c, Brn3c | |||
|
Domain Architecture |
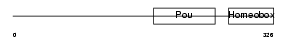
|
|||||
| Description | POU domain, class 4, transcription factor 3 (Brain-specific homeobox/POU domain protein 3C) (Brn-3C). | |||||
|
TAOK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | -0.003110 (rank : 119) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1172 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UL54, O94957, Q6UW73, Q7LC09, Q9NSW2 | Gene names | TAOK2, KIAA0881, MAP3K17, PSK, PSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2) (Prostate-derived STE20-like kinase 1) (PSK-1) (Kinase from chicken homolog C) (hKFC-C). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | -0.001642 (rank : 117) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.022350 (rank : 51) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.019679 (rank : 60) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.006392 (rank : 99) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.020666 (rank : 54) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
IRX5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.004400 (rank : 105) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |
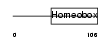
|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
KPCD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | -0.003673 (rank : 120) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94806 | Gene names | PRKD3, EPK2, PRKCN | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D3 (EC 2.7.11.13) (Protein kinase C nu type) (nPKC-nu) (Protein kinase EPK2). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.000257 (rank : 115) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.003865 (rank : 108) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.003782 (rank : 109) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.009522 (rank : 92) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
SIA7D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.002984 (rank : 111) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4F1, Q9NWU6, Q9UKU1, Q9ULB9, Q9Y3G3, Q9Y3G4 | Gene names | ST6GALNAC4, SIAT3C, SIAT7D | |||
|
Domain Architecture |
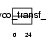
|
|||||
| Description | Alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3-N-acetyl- galactosaminide alpha-2,6-sialyltransferase (EC 2.4.99.7) (NeuAc- alpha-2,3-Gal-beta-1,3-GalNAc-alpha-2,6-sialyltransferase) (ST6GalNAc IV) (Sialyltransferase 7D) (Sialyltransferase 3C). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.008183 (rank : 96) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.012942 (rank : 75) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.014617 (rank : 70) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
STX11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.003019 (rank : 110) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D3G5 | Gene names | Stx11 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-11. | |||||
|
TBX3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
TBX3_MOUSE
|
||||||
| NC score | 0.992510 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70324, Q60708, Q8BWH7 | Gene names | Tbx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
TBX2_MOUSE
|
||||||
| NC score | 0.983041 (rank : 3) | θ value | 2.19465e-147 (rank : 4) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60707 | Gene names | Tbx2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TBX2_HUMAN
|
||||||
| NC score | 0.978805 (rank : 4) | θ value | 1.91815e-151 (rank : 3) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TBX20_MOUSE
|
||||||
| NC score | 0.968219 (rank : 5) | θ value | 4.10295e-61 (rank : 9) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ES03, Q9ESX1 | Gene names | Tbx20, Tbx12 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX20 (T-box protein 20). | |||||
|
TBX1_HUMAN
|
||||||
| NC score | 0.962020 (rank : 6) | θ value | 6.13105e-57 (rank : 11) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43435, O43436, Q96RJ2 | Gene names | TBX1 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX1 (T-box protein 1) (Testis-specific T- box protein). | |||||
|
TBX1_MOUSE
|
||||||
| NC score | 0.961830 (rank : 7) | θ value | 1.61343e-57 (rank : 10) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70323, Q60706, Q99MP0, Q99P22 | Gene names | Tbx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX1 (T-box protein 1) (Testis-specific T- box protein). | |||||
|
TBX10_HUMAN
|
||||||
| NC score | 0.959197 (rank : 8) | θ value | 2.32979e-56 (rank : 13) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75333, Q86XS3 | Gene names | TBX10, TBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10). | |||||
|
TBX6_HUMAN
|
||||||
| NC score | 0.959130 (rank : 9) | θ value | 1.78386e-56 (rank : 12) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95947, Q8TAS4 | Gene names | TBX6 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
TBX10_MOUSE
|
||||||
| NC score | 0.958444 (rank : 10) | θ value | 6.77864e-56 (rank : 14) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q810F8, O54841, Q6QH75 | Gene names | Tbx10, Tbx13, Tbx7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10) (T-box protein 13) (MmTBX7). | |||||
|
TBX6_MOUSE
|
||||||
| NC score | 0.955743 (rank : 11) | θ value | 9.78833e-55 (rank : 17) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70327 | Gene names | Tbx6 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
TBX5_HUMAN
|
||||||
| NC score | 0.954144 (rank : 12) | θ value | 8.26713e-62 (rank : 8) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99593, O15301, Q9Y4I2 | Gene names | TBX5 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX5 (T-box protein 5). | |||||
|
TBX4_MOUSE
|
||||||
| NC score | 0.954109 (rank : 13) | θ value | 6.76295e-64 (rank : 5) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P70325, Q8R5F6 | Gene names | Tbx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
TBX4_HUMAN
|
||||||
| NC score | 0.953761 (rank : 14) | θ value | 9.76566e-63 (rank : 6) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P57082 | Gene names | TBX4 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
TBX5_MOUSE
|
||||||
| NC score | 0.953615 (rank : 15) | θ value | 2.17557e-62 (rank : 7) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70326, Q9R003 | Gene names | Tbx5 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX5 (T-box protein 5). | |||||
|
TBX22_HUMAN
|
||||||
| NC score | 0.948186 (rank : 16) | θ value | 5.20228e-48 (rank : 20) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y458, Q96LC0, Q9HBF1 | Gene names | TBX22, TBOX22 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX22 (T-box protein 22). | |||||
|
TBX22_MOUSE
|
||||||
| NC score | 0.947707 (rank : 17) | θ value | 6.7944e-48 (rank : 21) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K402 | Gene names | Tbx22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX22 (T-box protein 22). | |||||
|
TBX18_MOUSE
|
||||||
| NC score | 0.944507 (rank : 18) | θ value | 5.73848e-55 (rank : 16) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EPZ6 | Gene names | Tbx18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
TBX18_HUMAN
|
||||||
| NC score | 0.941214 (rank : 19) | θ value | 3.36421e-55 (rank : 15) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
TBX15_HUMAN
|
||||||
| NC score | 0.941107 (rank : 20) | θ value | 2.66562e-52 (rank : 18) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SF7, Q5T9S7 | Gene names | TBX15, TBX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15). | |||||
|
TBX15_MOUSE
|
||||||
| NC score | 0.940979 (rank : 21) | θ value | 1.01294e-51 (rank : 19) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
BRAC_MOUSE
|
||||||
| NC score | 0.937562 (rank : 22) | θ value | 2.18568e-46 (rank : 23) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20293 | Gene names | T | |||
|
Domain Architecture |

|
|||||
| Description | Brachyury protein (T protein). | |||||
|
BRAC_HUMAN
|
||||||
| NC score | 0.936549 (rank : 23) | θ value | 7.51208e-47 (rank : 22) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15178 | Gene names | T | |||
|
Domain Architecture |

|
|||||
| Description | Brachyury protein (T protein). | |||||
|
TBX19_HUMAN
|
||||||
| NC score | 0.933570 (rank : 24) | θ value | 8.04462e-41 (rank : 30) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60806 | Gene names | TBX19, TPIT | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
TBX19_MOUSE
|
||||||
| NC score | 0.930321 (rank : 25) | θ value | 8.04462e-41 (rank : 31) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99ME7 | Gene names | Tbx19, Tpit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
TBX21_MOUSE
|
||||||
| NC score | 0.929228 (rank : 26) | θ value | 7.77379e-44 (rank : 27) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKD8, Q9R0A6 | Gene names | Tbx21, Tbet, Tblym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
TBX21_HUMAN
|
||||||
| NC score | 0.926462 (rank : 27) | θ value | 3.48949e-44 (rank : 26) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
EOMES_HUMAN
|
||||||
| NC score | 0.924568 (rank : 28) | θ value | 6.58091e-43 (rank : 28) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |

|
|||||
| Description | Eomesodermin homolog. | |||||
|
TBR1_MOUSE
|
||||||
| NC score | 0.923961 (rank : 29) | θ value | 1.85029e-45 (rank : 25) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q64336 | Gene names | Tbr1 | |||
|
Domain Architecture |

|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
TBR1_HUMAN
|
||||||
| NC score | 0.923934 (rank : 30) | θ value | 1.85029e-45 (rank : 24) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16650, Q53TH0 | Gene names | TBR1 | |||
|
Domain Architecture |

|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
EOMES_MOUSE
|
||||||
| NC score | 0.922929 (rank : 31) | θ value | 6.58091e-43 (rank : 29) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54839, Q9QYG7 | Gene names | Eomes, Tbr2 | |||
|
Domain Architecture |

|
|||||
| Description | Eomesodermin homolog. | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.053534 (rank : 32) | θ value | 0.00665767 (rank : 34) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.052541 (rank : 33) | θ value | 0.0252991 (rank : 35) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.052059 (rank : 34) | θ value | 0.0431538 (rank : 37) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.042453 (rank : 35) | θ value | 0.0431538 (rank : 36) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.040210 (rank : 36) | θ value | 0.00175202 (rank : 33) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.038287 (rank : 37) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.036573 (rank : 38) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.035228 (rank : 39) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.034075 (rank : 40) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.032993 (rank : 41) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.031756 (rank : 42) | θ value | 0.0431538 (rank : 38) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.031420 (rank : 43) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.028287 (rank : 44) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.025680 (rank : 45) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.024813 (rank : 46) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.024109 (rank : 47) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.023513 (rank : 48) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.023028 (rank : 49) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.022815 (rank : 50) | θ value | 9.29e-05 (rank : 32) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.022350 (rank : 51) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.021578 (rank : 52) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.021242 (rank : 53) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.020666 (rank : 54) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.020526 (rank : 55) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.020401 (rank : 56) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.020401 (rank : 57) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.020192 (rank : 58) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
SCG1_HUMAN
|
||||||
| NC score | 0.020020 (rank : 59) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.019679 (rank : 60) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.019091 (rank : 61) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
TULP1_HUMAN
|
||||||
| NC score | 0.018861 (rank : 62) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.018540 (rank : 63) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.018269 (rank : 64) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.017983 (rank : 65) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.017641 (rank : 66) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
GCM1_HUMAN
|
||||||
| NC score | 0.016986 (rank : 67) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NP62, Q5T0X0, Q99468, Q9P1X3 | Gene names | GCM1, GCMA | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (hGCMa). | |||||
|
TACC1_HUMAN
|
||||||
| NC score | 0.015701 (rank : 68) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.015531 (rank : 69) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.014617 (rank : 70) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.014435 (rank : 71) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.013356 (rank : 72) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.013310 (rank : 73) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.013119 (rank : 74) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.012942 (rank : 75) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.012684 (rank : 76) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.012208 (rank : 77) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.011754 (rank : 78) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
DIP2C_HUMAN
|
||||||
| NC score | 0.011297 (rank : 79) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2E4 | Gene names | DIP2C, KIAA0934 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog C. | |||||
|
HNRPF_MOUSE
|
||||||
| NC score | 0.011274 (rank : 80) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2X1, Q5FWK2, Q8BVU8, Q8K2U9, Q8R0E7 | Gene names | Hnrpf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein F (hnRNP F). | |||||
|
HNRPF_HUMAN
|
||||||
| NC score | 0.011244 (rank : 81) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52597, Q5T0N2, Q96AU2 | Gene names | HNRPF | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein F (hnRNP F) (Nucleolin-like protein mcs94-1). | |||||
|
NIPA_MOUSE
|
||||||
| NC score | 0.011167 (rank : 82) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.010956 (rank : 83) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.010946 (rank : 84) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
BIN1_MOUSE
|
||||||
| NC score | 0.010873 (rank : 85) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.010703 (rank : 86) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.010570 (rank : 87) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TLE4_HUMAN
|
||||||
| NC score | 0.010194 (rank : 88) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
TLE4_MOUSE
|
||||||
| NC score | 0.009880 (rank : 89) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
SAFB2_MOUSE
|
||||||
| NC score | 0.009867 (rank : 90) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
K1196_HUMAN
|
||||||
| NC score | 0.009564 (rank : 91) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.009522 (rank : 92) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RNF12_MOUSE
|
||||||
| NC score | 0.008428 (rank : 93) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.008387 (rank : 94) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.008347 (rank : 95) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.008183 (rank : 96) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
PO2F1_MOUSE
|
||||||
| NC score | 0.007688 (rank : 97) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P25425, Q61994, Q63891, Q6Y681, Q7TSD0, Q8BT04, Q8K570, Q99JH0, Q9WTZ4, Q9WTZ5, Q9WTZ6 | Gene names | Pou2f1, Oct-1, Otf-1, Otf1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.007045 (rank : 98) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.006392 (rank : 99) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
TAF1_HUMAN
|
||||||
| NC score | 0.006018 (rank : 100) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
OAS3_HUMAN
|
||||||
| NC score | 0.005799 (rank : 101) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
|
SH2D3_HUMAN
|
||||||
| NC score | 0.005509 (rank : 102) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.004901 (rank : 103) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.004673 (rank : 104) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
IRX5_HUMAN
|
||||||
| NC score | 0.004400 (rank : 105) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.004256 (rank : 106) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
S27A1_HUMAN
|
||||||
| NC score | 0.004017 (rank : 107) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PCB7 | Gene names | SLC27A1, ACSVL5, FATP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Long-chain fatty acid transport protein 1 (EC 6.2.1.-) (Fatty acid transport protein 1) (FATP-1) (Solute carrier family 27 member 1). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.003865 (rank : 108) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.003782 (rank : 109) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
STX11_MOUSE
|
||||||
| NC score | 0.003019 (rank : 110) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D3G5 | Gene names | Stx11 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-11. | |||||
|
SIA7D_HUMAN
|
||||||
| NC score | 0.002984 (rank : 111) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4F1, Q9NWU6, Q9UKU1, Q9ULB9, Q9Y3G3, Q9Y3G4 | Gene names | ST6GALNAC4, SIAT3C, SIAT7D | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3-N-acetyl- galactosaminide alpha-2,6-sialyltransferase (EC 2.4.99.7) (NeuAc- alpha-2,3-Gal-beta-1,3-GalNAc-alpha-2,6-sialyltransferase) (ST6GalNAc IV) (Sialyltransferase 7D) (Sialyltransferase 3C). | |||||
|
PO4F3_MOUSE
|
||||||
| NC score | 0.002831 (rank : 112) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63955 | Gene names | Pou4f3, Brn-3c, Brn3c | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 3 (Brain-specific homeobox/POU domain protein 3C) (Brn-3C). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.001215 (rank : 113) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.000833 (rank : 114) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.000257 (rank : 115) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.000242 (rank : 116) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | -0.001642 (rank : 117) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
TAOK2_MOUSE
|
||||||
| NC score | -0.001875 (rank : 118) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
TAOK2_HUMAN
|
||||||
| NC score | -0.003110 (rank : 119) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1172 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UL54, O94957, Q6UW73, Q7LC09, Q9NSW2 | Gene names | TAOK2, KIAA0881, MAP3K17, PSK, PSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2) (Prostate-derived STE20-like kinase 1) (PSK-1) (Kinase from chicken homolog C) (hKFC-C). | |||||
|
KPCD3_HUMAN
|
||||||
| NC score | -0.003673 (rank : 120) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94806 | Gene names | PRKD3, EPK2, PRKCN | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D3 (EC 2.7.11.13) (Protein kinase C nu type) (nPKC-nu) (Protein kinase EPK2). | |||||
|
ADA2B_HUMAN
|
||||||
| NC score | -0.003751 (rank : 121) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 881 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18089, Q4TUH9, Q53RF2, Q9BZK0 | Gene names | ADRA2B | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2B adrenergic receptor (Alpha-2B adrenoceptor) (Alpha-2B adrenoreceptor) (Alpha-2 adrenergic receptor subtype C2). | |||||