Please be patient as the page loads
|
SIM1_MOUSE
|
||||||
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SIM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998753 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.988506 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.987324 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 1.65806e-78 (rank : 5) | NC score | 0.937034 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 2.1655e-78 (rank : 6) | NC score | 0.938104 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 4.99229e-75 (rank : 7) | NC score | 0.947077 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | 1.04096e-72 (rank : 8) | NC score | 0.945290 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 1.7756e-72 (rank : 9) | NC score | 0.932898 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 5.1662e-72 (rank : 10) | NC score | 0.929965 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 3.35642e-63 (rank : 11) | NC score | 0.920668 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 9.76566e-63 (rank : 12) | NC score | 0.921283 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 1.6813e-30 (rank : 13) | NC score | 0.797677 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 14) | NC score | 0.799101 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 1.08979e-29 (rank : 15) | NC score | 0.834025 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 5.97985e-28 (rank : 16) | NC score | 0.834601 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 1.80048e-24 (rank : 17) | NC score | 0.752354 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 18) | NC score | 0.750912 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 19) | NC score | 0.763848 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 1.99067e-23 (rank : 20) | NC score | 0.761162 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 21) | NC score | 0.740924 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 22) | NC score | 0.728266 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 23) | NC score | 0.692197 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 24) | NC score | 0.739351 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 25) | NC score | 0.676622 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 26) | NC score | 0.731920 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 27) | NC score | 0.641130 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 28) | NC score | 0.637614 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 29) | NC score | 0.565755 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 30) | NC score | 0.563301 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 31) | NC score | 0.561427 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 32) | NC score | 0.524071 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 33) | NC score | 0.253722 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 34) | NC score | 0.231877 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 35) | NC score | 0.261653 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
M3K3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 36) | NC score | -0.000778 (rank : 78) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61084 | Gene names | Map3k3, Mekk3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 3 (EC 2.7.11.25) (MAPK/ERK kinase kinase 3) (MEK kinase 3) (MEKK 3). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.024971 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.007398 (rank : 72) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.032969 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
AREG_MOUSE
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.040441 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31955 | Gene names | Areg, Sdgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amphiregulin precursor (AR) (Schwannoma-derived growth factor) (SDGF). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.237752 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.233710 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.231367 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.019740 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.027020 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.019247 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
3BHS5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.007556 (rank : 71) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61694, Q91X27 | Gene names | Hsd3b5 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 5 (3 beta-hydroxysteroid dehydrogenase type V) (3Beta-HSD V) (NADPH-dependent 3-beta-hydroxy- delta(5)-steroid dehydrogenase) (EC 1.1.1.-) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase). | |||||
|
ZN174_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | -0.002198 (rank : 79) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15697, Q9BQ34 | Gene names | ZNF174, ZSCAN8 | |||
|
Domain Architecture |
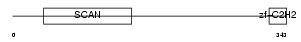
|
|||||
| Description | Zinc finger protein 174 (AW-1) (Zinc finger and SCAN domain-containing protein 8). | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.020477 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
PDE4C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.005947 (rank : 74) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.011300 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.099057 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.102816 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.109242 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.107336 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.002002 (rank : 76) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
HELI_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | -0.003181 (rank : 80) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P81183 | Gene names | Ikzf2, Helios, Zfpn1a2, Znfn1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Helios (IKAROS family zinc finger protein 2). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.011286 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PITM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.009303 (rank : 70) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
TNAP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.015036 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61333, Q922G1 | Gene names | Tnfaip2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 2 (Primary response gene B94 protein). | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.006954 (rank : 73) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.001632 (rank : 77) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
PRIC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.004431 (rank : 75) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MT3, Q71QF8, Q96N00 | Gene names | PRICKLE1, RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 1 (REST/NRSF-interacting LIM domain protein 1). | |||||
|
RBNS5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.011904 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
TAF1C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.016459 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PDZ2, P97359, Q3U3B6, Q8BN54 | Gene names | Taf1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor, RNA polymerase I, subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 95 kDa) (TAFI95). | |||||
|
HES5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.070877 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | -0.004448 (rank : 81) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
PP1RB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.011891 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60927 | Gene names | PPP1R11, HCGV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 11 (Protein phosphatase inhibitor 3) (Hemochromatosis candidate protein V) (HCG V) (HCG-V). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.022915 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.068444 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.070005 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.063885 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.063902 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.069764 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.068395 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.064414 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.064370 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.064106 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.064077 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.061810 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.057788 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.998753 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.988506 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.987324 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.947077 (rank : 5) | θ value | 4.99229e-75 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.945290 (rank : 6) | θ value | 1.04096e-72 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.938104 (rank : 7) | θ value | 2.1655e-78 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.937034 (rank : 8) | θ value | 1.65806e-78 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.932898 (rank : 9) | θ value | 1.7756e-72 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.929965 (rank : 10) | θ value | 5.1662e-72 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.921283 (rank : 11) | θ value | 9.76566e-63 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.920668 (rank : 12) | θ value | 3.35642e-63 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.834601 (rank : 13) | θ value | 5.97985e-28 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.834025 (rank : 14) | θ value | 1.08979e-29 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.799101 (rank : 15) | θ value | 4.89182e-30 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.797677 (rank : 16) | θ value | 1.6813e-30 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.763848 (rank : 17) | θ value | 1.16704e-23 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.761162 (rank : 18) | θ value | 1.99067e-23 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.752354 (rank : 19) | θ value | 1.80048e-24 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.750912 (rank : 20) | θ value | 5.23862e-24 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.740924 (rank : 21) | θ value | 1.29031e-22 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.739351 (rank : 22) | θ value | 2.27762e-19 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.731920 (rank : 23) | θ value | 1.13037e-18 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.728266 (rank : 24) | θ value | 1.6852e-22 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.692197 (rank : 25) | θ value | 7.82807e-20 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.676622 (rank : 26) | θ value | 8.65492e-19 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.641130 (rank : 27) | θ value | 3.28887e-18 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.637614 (rank : 28) | θ value | 7.32683e-18 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.565755 (rank : 29) | θ value | 2.43908e-13 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.563301 (rank : 30) | θ value | 1.2105e-12 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.561427 (rank : 31) | θ value | 4.30538e-10 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.524071 (rank : 32) | θ value | 4.0297e-08 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.261653 (rank : 33) | θ value | 0.0431538 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.253722 (rank : 34) | θ value | 0.00134147 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.237752 (rank : 35) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.233710 (rank : 36) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.231877 (rank : 37) | θ value | 0.00869519 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.231367 (rank : 38) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.109242 (rank : 39) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.107336 (rank : 40) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.102816 (rank : 41) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.099057 (rank : 42) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.070877 (rank : 43) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.070005 (rank : 44) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.069764 (rank : 45) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.068444 (rank : 46) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.068395 (rank : 47) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.064414 (rank : 48) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.064370 (rank : 49) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.064106 (rank : 50) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.064077 (rank : 51) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.063902 (rank : 52) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.063885 (rank : 53) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.061810 (rank : 54) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.057788 (rank : 55) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
AREG_MOUSE
|
||||||
| NC score | 0.040441 (rank : 56) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31955 | Gene names | Areg, Sdgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amphiregulin precursor (AR) (Schwannoma-derived growth factor) (SDGF). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.032969 (rank : 57) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.027020 (rank : 58) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.024971 (rank : 59) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.022915 (rank : 60) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.020477 (rank : 61) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.019740 (rank : 62) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.019247 (rank : 63) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TAF1C_MOUSE
|
||||||
| NC score | 0.016459 (rank : 64) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PDZ2, P97359, Q3U3B6, Q8BN54 | Gene names | Taf1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor, RNA polymerase I, subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 95 kDa) (TAFI95). | |||||
|
TNAP2_MOUSE
|
||||||
| NC score | 0.015036 (rank : 65) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61333, Q922G1 | Gene names | Tnfaip2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 2 (Primary response gene B94 protein). | |||||
|
RBNS5_HUMAN
|
||||||
| NC score | 0.011904 (rank : 66) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
PP1RB_HUMAN
|
||||||
| NC score | 0.011891 (rank : 67) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60927 | Gene names | PPP1R11, HCGV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 11 (Protein phosphatase inhibitor 3) (Hemochromatosis candidate protein V) (HCG V) (HCG-V). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.011300 (rank : 68) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.011286 (rank : 69) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PITM2_HUMAN
|
||||||
| NC score | 0.009303 (rank : 70) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
3BHS5_MOUSE
|
||||||
| NC score | 0.007556 (rank : 71) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61694, Q91X27 | Gene names | Hsd3b5 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 5 (3 beta-hydroxysteroid dehydrogenase type V) (3Beta-HSD V) (NADPH-dependent 3-beta-hydroxy- delta(5)-steroid dehydrogenase) (EC 1.1.1.-) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.007398 (rank : 72) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.006954 (rank : 73) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
PDE4C_HUMAN
|
||||||
| NC score | 0.005947 (rank : 74) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PRIC1_HUMAN
|
||||||
| NC score | 0.004431 (rank : 75) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MT3, Q71QF8, Q96N00 | Gene names | PRICKLE1, RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 1 (REST/NRSF-interacting LIM domain protein 1). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.002002 (rank : 76) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.001632 (rank : 77) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
M3K3_MOUSE
|
||||||
| NC score | -0.000778 (rank : 78) | θ value | 0.21417 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61084 | Gene names | Map3k3, Mekk3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 3 (EC 2.7.11.25) (MAPK/ERK kinase kinase 3) (MEK kinase 3) (MEKK 3). | |||||
|
ZN174_HUMAN
|
||||||
| NC score | -0.002198 (rank : 79) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15697, Q9BQ34 | Gene names | ZNF174, ZSCAN8 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 174 (AW-1) (Zinc finger and SCAN domain-containing protein 8). | |||||
|
HELI_MOUSE
|
||||||
| NC score | -0.003181 (rank : 80) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P81183 | Gene names | Ikzf2, Helios, Zfpn1a2, Znfn1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Helios (IKAROS family zinc finger protein 2). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | -0.004448 (rank : 81) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||