Please be patient as the page loads
|
HES4_HUMAN
|
||||||
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HES4_HUMAN
|
||||||
| θ value | 1.00825e-67 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
HES1_HUMAN
|
||||||
| θ value | 1.56274e-52 (rank : 2) | NC score | 0.979292 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | 1.56274e-52 (rank : 3) | NC score | 0.977661 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES2_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 4) | NC score | 0.925658 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES2_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 5) | NC score | 0.916258 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 6) | NC score | 0.813659 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 7) | NC score | 0.813024 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 8) | NC score | 0.796417 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HES6_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 9) | NC score | 0.818421 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 10) | NC score | 0.798989 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HES6_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 11) | NC score | 0.809238 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
HES7_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 12) | NC score | 0.747464 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 13) | NC score | 0.750303 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES5_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 14) | NC score | 0.849466 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 15) | NC score | 0.613807 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 16) | NC score | 0.607797 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 17) | NC score | 0.597139 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 18) | NC score | 0.609596 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
HES3_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 19) | NC score | 0.714274 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
MITF_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.213779 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.209548 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.214406 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MAD1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.133884 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.176583 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.028407 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.153142 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.155603 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
USF2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.180311 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.183238 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.025132 (rank : 65) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.186360 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.098277 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.098514 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.117909 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.158414 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.072986 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.072869 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
MAD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.112237 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
USF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.190833 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.190910 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MYF6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.071754 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |
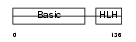
|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.071247 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |
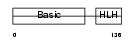
|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.004350 (rank : 69) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.176159 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.178321 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.008170 (rank : 67) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.086891 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYF5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.050235 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |
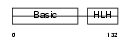
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.049871 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |
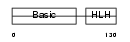
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYOD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.081039 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYOD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.081722 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
CO1A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.004479 (rank : 68) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | -0.000456 (rank : 70) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYOG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.076367 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |
|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.075920 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |
|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.094545 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.096194 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
MMAC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.012169 (rank : 66) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4U1, Q5T157, Q9BRQ7 | Gene names | MMACHC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methylmalonic aciduria and homocystinuria type C protein. | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.068688 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.067138 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.072754 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.058355 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.071967 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.073893 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.133155 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.135424 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.087532 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.086741 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.055530 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.055333 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.00825e-67 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.979292 (rank : 2) | θ value | 1.56274e-52 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.977661 (rank : 3) | θ value | 1.56274e-52 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES2_MOUSE
|
||||||
| NC score | 0.925658 (rank : 4) | θ value | 4.43474e-23 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES2_HUMAN
|
||||||
| NC score | 0.916258 (rank : 5) | θ value | 8.36355e-22 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.849466 (rank : 6) | θ value | 4.30538e-10 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HES6_HUMAN
|
||||||
| NC score | 0.818421 (rank : 7) | θ value | 9.90251e-15 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.813659 (rank : 8) | θ value | 8.10077e-17 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.813024 (rank : 9) | θ value | 8.10077e-17 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HES6_MOUSE
|
||||||
| NC score | 0.809238 (rank : 10) | θ value | 2.88119e-14 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.798989 (rank : 11) | θ value | 9.90251e-15 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.796417 (rank : 12) | θ value | 1.52774e-15 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HES7_HUMAN
|
||||||
| NC score | 0.750303 (rank : 13) | θ value | 6.00763e-12 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| NC score | 0.747464 (rank : 14) | θ value | 3.52202e-12 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES3_MOUSE
|
||||||
| NC score | 0.714274 (rank : 15) | θ value | 1.17247e-07 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.613807 (rank : 16) | θ value | 4.76016e-09 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.609596 (rank : 17) | θ value | 8.97725e-08 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.607797 (rank : 18) | θ value | 6.21693e-09 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.597139 (rank : 19) | θ value | 8.11959e-09 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.214406 (rank : 20) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.213779 (rank : 21) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.209548 (rank : 22) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.190910 (rank : 23) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.190833 (rank : 24) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.186360 (rank : 25) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.183238 (rank : 26) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.180311 (rank : 27) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.178321 (rank : 28) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.176583 (rank : 29) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.176159 (rank : 30) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.158414 (rank : 31) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.155603 (rank : 32) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.153142 (rank : 33) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.135424 (rank : 34) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.133884 (rank : 35) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.133155 (rank : 36) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.117909 (rank : 37) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.112237 (rank : 38) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.098514 (rank : 39) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.098277 (rank : 40) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.096194 (rank : 41) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.094545 (rank : 42) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.087532 (rank : 43) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.086891 (rank : 44) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.086741 (rank : 45) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MYOD1_MOUSE
|
||||||
| NC score | 0.081722 (rank : 46) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MYOD1_HUMAN
|
||||||
| NC score | 0.081039 (rank : 47) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYOG_HUMAN
|
||||||
| NC score | 0.076367 (rank : 48) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| NC score | 0.075920 (rank : 49) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.073893 (rank : 50) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.072986 (rank : 51) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.072869 (rank : 52) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.072754 (rank : 53) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.071967 (rank : 54) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MYF6_HUMAN
|
||||||
| NC score | 0.071754 (rank : 55) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| NC score | 0.071247 (rank : 56) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.068688 (rank : 57) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.067138 (rank : 58) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.058355 (rank : 59) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.055530 (rank : 60) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.055333 (rank : 61) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
MYF5_HUMAN
|
||||||
| NC score | 0.050235 (rank : 62) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF5_MOUSE
|
||||||
| NC score | 0.049871 (rank : 63) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.028407 (rank : 64) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.025132 (rank : 65) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
MMAC_HUMAN
|
||||||
| NC score | 0.012169 (rank : 66) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4U1, Q5T157, Q9BRQ7 | Gene names | MMACHC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methylmalonic aciduria and homocystinuria type C protein. | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.008170 (rank : 67) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
CO1A2_MOUSE
|
||||||
| NC score | 0.004479 (rank : 68) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.004350 (rank : 69) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | -0.000456 (rank : 70) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||