Please be patient as the page loads
|
EXOC3_HUMAN
|
||||||
| SwissProt Accessions | O60645, Q8TEN6, Q8WUW0, Q96DI4 | Gene names | EXOC3, SEC6, SEC6L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EXOC3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O60645, Q8TEN6, Q8WUW0, Q96DI4 | Gene names | EXOC3, SEC6, SEC6L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
|
EXOC3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992067 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6KAR6 | Gene names | Exoc3, Sec6l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
|
TNAP2_MOUSE
|
||||||
| θ value | 1.24399e-33 (rank : 3) | NC score | 0.710449 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61333, Q922G1 | Gene names | Tnfaip2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 2 (Primary response gene B94 protein). | |||||
|
TNAP2_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 4) | NC score | 0.703605 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03169, Q86VI0 | Gene names | TNFAIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 2 (Primary response gene B94 protein). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 5) | NC score | 0.095926 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 6) | NC score | 0.096287 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
K1C14_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 7) | NC score | 0.040773 (rank : 123) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |
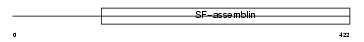
|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
TRIM3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 8) | NC score | 0.066367 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9R1R2, Q3UMU5 | Gene names | Trim3, Hac1, Rnf22 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 3 (RING finger protein 22) (RING finger protein HAC1). | |||||
|
TRIM3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.064301 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75382, Q9C038, Q9C039 | Gene names | TRIM3, BERP, RNF22, RNF97 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 3 (RING finger protein 22) (Brain- expressed RING finger protein) (RING finger protein 97). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.083019 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.073913 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.073407 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.077523 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
GPC4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.050815 (rank : 111) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51655, Q923C8 | Gene names | Gpc4 | |||
|
Domain Architecture |

|
|||||
| Description | Glypican-4 precursor (K-glypican). | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.066448 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
ECM29_MOUSE
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.053212 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PDI5, Q6ZQC9, Q8BSW7, Q8CAH0, Q8R3M6 | Gene names | Ecm29, Kiaa0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.059127 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.062905 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.058509 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.070331 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.070469 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
GPC4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.043837 (rank : 121) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75487, Q6ZMA6, Q96L43, Q9NU08, Q9UJN1, Q9UPD9 | Gene names | GPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Glypican-4 precursor (K-glypican). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.075415 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
PERI_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.045771 (rank : 119) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41219, Q8N577 | Gene names | PRPH | |||
|
Domain Architecture |
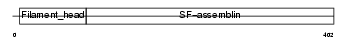
|
|||||
| Description | Peripherin. | |||||
|
PERI_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.045664 (rank : 120) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15331, O35688, O35689 | Gene names | Prph, Prph1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.072853 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.063598 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.067534 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.013828 (rank : 141) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.058403 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.057987 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.070762 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.058281 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
K1C12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.036411 (rank : 124) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99456 | Gene names | KRT12 | |||
|
Domain Architecture |
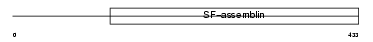
|
|||||
| Description | Keratin, type I cytoskeletal 12 (Cytokeratin-12) (CK-12) (Keratin-12) (K12). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.071227 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.012172 (rank : 143) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.009798 (rank : 145) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.061275 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.067567 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.063697 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
DIAP3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.030827 (rank : 130) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z207 | Gene names | Diaph3, Diap3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3) (mDIA2) (p134mDIA2). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.062165 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
EP15_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.068305 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.017194 (rank : 137) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.065887 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.026175 (rank : 132) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
TRIM6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.019504 (rank : 136) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGE7, Q99PQ6 | Gene names | Trim6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 6. | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.059846 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.055403 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.059688 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.059785 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.064192 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LIAS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.032173 (rank : 129) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43766, Q8IV62 | Gene names | LIAS, LAS | |||
|
Domain Architecture |
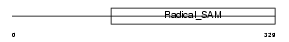
|
|||||
| Description | Lipoic acid synthetase, mitochondrial precursor (Lip-syn) (Lipoate synthase). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.054457 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.054814 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NFL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.034402 (rank : 125) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
PARP9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.022032 (rank : 134) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CAS9, Q99LF9 | Gene names | Parp9, Bal | |||
|
Domain Architecture |
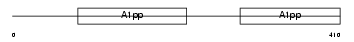
|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein homolog). | |||||
|
PPIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.014422 (rank : 140) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43586, O43585, O95657 | Gene names | PSTPIP1, CD2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-serine-threonine phosphatase-interacting protein 1 (PEST phosphatase-interacting protein 1) (CD2-binding protein 1) (H-PIP). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.062879 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RFIP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.050842 (rank : 109) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BQP8, Q80T87 | Gene names | Rab11fip4, Kiaa1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
TUFT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.053558 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
CHMP6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.021256 (rank : 135) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96FZ7, Q53FU4, Q9HAE8 | Gene names | CHMP6, VPS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6) (Vacuolar protein sorting-associated protein 20) (hVps20). | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.059475 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
FGD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.012693 (rank : 142) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
GRPE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.026019 (rank : 133) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88396, Q3ULH8, Q9CQ97 | Gene names | Grpel2, Mt-grpel2 | |||
|
Domain Architecture |
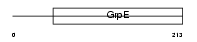
|
|||||
| Description | GrpE protein homolog 2, mitochondrial precursor (Mt-GrpE#2). | |||||
|
PROD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.017057 (rank : 138) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WU79 | Gene names | Prodh, Pro1 | |||
|
Domain Architecture |

|
|||||
| Description | Proline oxidase, mitochondrial precursor (EC 1.5.3.-) (Proline dehydrogenase). | |||||
|
SAMH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.027931 (rank : 131) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60710, Q91VK8 | Gene names | Samhd1, Mg11 | |||
|
Domain Architecture |
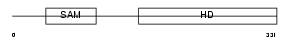
|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Interferon-gamma- inducible protein Mg11). | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.016811 (rank : 139) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
TS101_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.041691 (rank : 122) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |
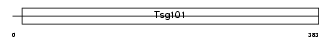
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.009868 (rank : 144) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
IF35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.034174 (rank : 126) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
IF35_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.032190 (rank : 128) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.053287 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NFL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.033709 (rank : 127) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.054883 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.050606 (rank : 114) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.052647 (rank : 82) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052922 (rank : 79) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CCD39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.057205 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.050772 (rank : 112) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.051329 (rank : 100) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
CE290_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.052106 (rank : 87) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.052651 (rank : 81) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.050663 (rank : 113) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CF060_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.052322 (rank : 86) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CI093_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.052884 (rank : 80) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CING_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.054667 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CING_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.051977 (rank : 90) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.051234 (rank : 102) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CN145_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.050378 (rank : 115) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
CP135_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.055180 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP135_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.052078 (rank : 89) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP250_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.051211 (rank : 103) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CP250_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.055174 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.056474 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.051877 (rank : 92) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.051820 (rank : 93) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
DMD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.054483 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.058020 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
EEA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.057188 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
EP15R_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.053050 (rank : 75) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.052949 (rank : 77) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.052103 (rank : 88) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.055688 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.050111 (rank : 117) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.051498 (rank : 98) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.055393 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.054499 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.052385 (rank : 85) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051541 (rank : 97) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.050823 (rank : 110) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
K0555_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.050856 (rank : 108) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.059076 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.056416 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054228 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.051547 (rank : 96) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050993 (rank : 107) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.051728 (rank : 94) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051616 (rank : 95) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.051192 (rank : 104) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.051930 (rank : 91) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.051141 (rank : 106) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.050362 (rank : 116) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.051285 (rank : 101) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.053541 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.050099 (rank : 118) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.051167 (rank : 105) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.052932 (rank : 78) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.055393 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052523 (rank : 83) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.051470 (rank : 99) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.058113 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
REST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.053028 (rank : 76) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.057405 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.056968 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.055996 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.062857 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.063270 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.056653 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.057776 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.054487 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.058400 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.052390 (rank : 84) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.053684 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.057082 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
EXOC3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O60645, Q8TEN6, Q8WUW0, Q96DI4 | Gene names | EXOC3, SEC6, SEC6L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
|
EXOC3_MOUSE
|
||||||
| NC score | 0.992067 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6KAR6 | Gene names | Exoc3, Sec6l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
|
TNAP2_MOUSE
|
||||||
| NC score | 0.710449 (rank : 3) | θ value | 1.24399e-33 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61333, Q922G1 | Gene names | Tnfaip2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 2 (Primary response gene B94 protein). | |||||
|
TNAP2_HUMAN
|
||||||
| NC score | 0.703605 (rank : 4) | θ value | 1.98606e-31 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03169, Q86VI0 | Gene names | TNFAIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 2 (Primary response gene B94 protein). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.096287 (rank : 5) | θ value | 0.000158464 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.095926 (rank : 6) | θ value | 0.000158464 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.083019 (rank : 7) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.077523 (rank : 8) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.075415 (rank : 9) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.073913 (rank : 10) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.073407 (rank : 11) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.072853 (rank : 12) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.071227 (rank : 13) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.070762 (rank : 14) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.070469 (rank : 15) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.070331 (rank : 16) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.068305 (rank : 17) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.067567 (rank : 18) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.067534 (rank : 19) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.066448 (rank : 20) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
TRIM3_MOUSE
|
||||||
| NC score | 0.066367 (rank : 21) | θ value | 0.0252991 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9R1R2, Q3UMU5 | Gene names | Trim3, Hac1, Rnf22 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 3 (RING finger protein 22) (RING finger protein HAC1). | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.065887 (rank : 22) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
TRIM3_HUMAN
|
||||||
| NC score | 0.064301 (rank : 23) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75382, Q9C038, Q9C039 | Gene names | TRIM3, BERP, RNF22, RNF97 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 3 (RING finger protein 22) (Brain- expressed RING finger protein) (RING finger protein 97). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.064192 (rank : 24) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.063697 (rank : 25) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.063598 (rank : 26) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.063270 (rank : 27) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.062905 (rank : 28) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.062879 (rank : 29) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.062857 (rank : 30) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.062165 (rank : 31) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.061275 (rank : 32) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.059846 (rank : 33) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.059785 (rank : 34) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.059688 (rank : 35) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.059475 (rank : 36) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.059127 (rank : 37) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.059076 (rank : 38) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.058509 (rank : 39) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.058403 (rank : 40) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.058400 (rank : 41) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.058281 (rank : 42) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.058113 (rank : 43) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.058020 (rank : 44) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.057987 (rank : 45) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.057776 (rank : 46) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.057405 (rank : 47) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.057205 (rank : 48) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.057188 (rank : 49) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.057082 (rank : 50) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.056968 (rank : 51) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.056653 (rank : 52) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.056474 (rank : 53) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.056416 (rank : 54) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.055996 (rank : 55) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.055688 (rank : 56) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.055403 (rank : 57) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.055393 (rank : 58) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.055393 (rank : 59) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.055180 (rank : 60) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.055174 (rank : 61) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.054883 (rank : 62) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.054814 (rank : 63) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.054667 (rank : 64) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.054499 (rank : 65) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.054487 (rank : 66) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.054483 (rank : 67) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.054457 (rank : 68) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.054228 (rank : 69) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.053684 (rank : 70) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
TUFT1_HUMAN
|
||||||
| NC score | 0.053558 (rank : 71) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.053541 (rank : 72) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.053287 (rank : 73) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
ECM29_MOUSE
|
||||||
| NC score | 0.053212 (rank : 74) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PDI5, Q6ZQC9, Q8BSW7, Q8CAH0, Q8R3M6 | Gene names | Ecm29, Kiaa0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.053050 (rank : 75) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.053028 (rank : 76) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.052949 (rank : 77) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.052932 (rank : 78) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.052922 (rank : 79) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.052884 (rank : 80) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.052651 (rank : 81) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.052647 (rank : 82) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.052523 (rank : 83) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.052390 (rank : 84) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.052385 (rank : 85) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.052322 (rank : 86) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.052106 (rank : 87) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.052103 (rank : 88) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.052078 (rank : 89) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.051977 (rank : 90) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.051930 (rank : 91) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.051877 (rank : 92) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.051820 (rank : 93) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.051728 (rank : 94) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.051616 (rank : 95) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.051547 (rank : 96) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.051541 (rank : 97) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.051498 (rank : 98) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.051470 (rank : 99) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.051329 (rank : 100) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.051285 (rank : 101) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.051234 (rank : 102) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.051211 (rank : 103) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.051192 (rank : 104) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.051167 (rank : 105) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.051141 (rank : 106) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.050993 (rank : 107) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.050856 (rank : 108) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
RFIP4_MOUSE
|
||||||
| NC score | 0.050842 (rank : 109) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BQP8, Q80T87 | Gene names | Rab11fip4, Kiaa1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.050823 (rank : 110) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
GPC4_MOUSE
|
||||||
| NC score | 0.050815 (rank : 111) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51655, Q923C8 | Gene names | Gpc4 | |||
|
Domain Architecture |

|
|||||
| Description | Glypican-4 precursor (K-glypican). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.050772 (rank : 112) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.050663 (rank : 113) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.050606 (rank : 114) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.050378 (rank : 115) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.050362 (rank : 116) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.050111 (rank : 117) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.050099 (rank : 118) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
PERI_HUMAN
|
||||||
| NC score | 0.045771 (rank : 119) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41219, Q8N577 | Gene names | PRPH | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
PERI_MOUSE
|
||||||
| NC score | 0.045664 (rank : 120) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15331, O35688, O35689 | Gene names | Prph, Prph1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
GPC4_HUMAN
|
||||||
| NC score | 0.043837 (rank : 121) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75487, Q6ZMA6, Q96L43, Q9NU08, Q9UJN1, Q9UPD9 | Gene names | GPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Glypican-4 precursor (K-glypican). | |||||
|
TS101_MOUSE
|
||||||
| NC score | 0.041691 (rank : 122) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
K1C14_HUMAN
|
||||||
| NC score | 0.040773 (rank : 123) | θ value | 0.00869519 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
K1C12_HUMAN
|
||||||
| NC score | 0.036411 (rank : 124) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99456 | Gene names | KRT12 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 12 (Cytokeratin-12) (CK-12) (Keratin-12) (K12). | |||||
|
NFL_HUMAN
|
||||||
| NC score | 0.034402 (rank : 125) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
IF35_HUMAN
|
||||||
| NC score | 0.034174 (rank : 126) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
NFL_MOUSE
|
||||||
| NC score | 0.033709 (rank : 127) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.032190 (rank : 128) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
LIAS_HUMAN
|
||||||
| NC score | 0.032173 (rank : 129) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43766, Q8IV62 | Gene names | LIAS, LAS | |||
|
Domain Architecture |

|
|||||
| Description | Lipoic acid synthetase, mitochondrial precursor (Lip-syn) (Lipoate synthase). | |||||
|
DIAP3_MOUSE
|
||||||
| NC score | 0.030827 (rank : 130) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z207 | Gene names | Diaph3, Diap3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3) (mDIA2) (p134mDIA2). | |||||
|
SAMH1_MOUSE
|
||||||
| NC score | 0.027931 (rank : 131) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60710, Q91VK8 | Gene names | Samhd1, Mg11 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Interferon-gamma- inducible protein Mg11). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.026175 (rank : 132) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
GRPE2_MOUSE
|
||||||
| NC score | 0.026019 (rank : 133) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88396, Q3ULH8, Q9CQ97 | Gene names | Grpel2, Mt-grpel2 | |||
|
Domain Architecture |

|
|||||
| Description | GrpE protein homolog 2, mitochondrial precursor (Mt-GrpE#2). | |||||
|
PARP9_MOUSE
|
||||||
| NC score | 0.022032 (rank : 134) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CAS9, Q99LF9 | Gene names | Parp9, Bal | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein homolog). | |||||
|
CHMP6_HUMAN
|
||||||
| NC score | 0.021256 (rank : 135) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96FZ7, Q53FU4, Q9HAE8 | Gene names | CHMP6, VPS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6) (Vacuolar protein sorting-associated protein 20) (hVps20). | |||||
|
TRIM6_MOUSE
|
||||||
| NC score | 0.019504 (rank : 136) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGE7, Q99PQ6 | Gene names | Trim6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 6. | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.017194 (rank : 137) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
PROD_MOUSE
|
||||||
| NC score | 0.017057 (rank : 138) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WU79 | Gene names | Prodh, Pro1 | |||
|
Domain Architecture |

|
|||||
| Description | Proline oxidase, mitochondrial precursor (EC 1.5.3.-) (Proline dehydrogenase). | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.016811 (rank : 139) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
PPIP1_HUMAN
|
||||||
| NC score | 0.014422 (rank : 140) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43586, O43585, O95657 | Gene names | PSTPIP1, CD2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-serine-threonine phosphatase-interacting protein 1 (PEST phosphatase-interacting protein 1) (CD2-binding protein 1) (H-PIP). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.013828 (rank : 141) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
FGD4_HUMAN
|
||||||
| NC score | 0.012693 (rank : 142) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.012172 (rank : 143) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.009868 (rank : 144) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.009798 (rank : 145) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||