Please be patient as the page loads
|
STON2_HUMAN
|
||||||
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STON2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
STON2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.973763 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
STON1_MOUSE
|
||||||
| θ value | 1.58745e-113 (rank : 3) | NC score | 0.904887 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
STON1_HUMAN
|
||||||
| θ value | 2.70778e-113 (rank : 4) | NC score | 0.906721 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6Q2, Q96JE3, Q9BYX3 | Gene names | STON1, SALF, SBLF, STN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
AP2M1_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 5) | NC score | 0.469648 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96CW1, P20172, P53679 | Gene names | AP2M1, CLAPM1, KIAA0109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (HA2 50 kDa subunit) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP2M1_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 6) | NC score | 0.469648 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84091, P20172, P53679 | Gene names | Ap2m1, Clapm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP1M1_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 7) | NC score | 0.460841 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BXS5 | Gene names | AP1M1, CLTNM | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M1_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 8) | NC score | 0.458140 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35585 | Gene names | Ap1m1, Cltnm | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M2_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 9) | NC score | 0.448017 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6Q5, Q9BSI8 | Gene names | AP1M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP1M2_MOUSE
|
||||||
| θ value | 9.26847e-13 (rank : 10) | NC score | 0.447466 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVP1, Q99LA4, Q9CWP7 | Gene names | Ap1m2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
ECM2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 11) | NC score | 0.040513 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94769 | Gene names | ECM2 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 2 precursor (Matrix glycoprotein SC1/ECM2). | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 12) | NC score | 0.070160 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 13) | NC score | 0.055764 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 14) | NC score | 0.061615 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 15) | NC score | 0.072474 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
AP4M1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 16) | NC score | 0.304589 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00189, Q9UHK9 | Gene names | AP4M1, MUARP2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.064068 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
EYA3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 18) | NC score | 0.036151 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.075374 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PALB2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.078085 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
BIRC2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.026517 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.038150 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.039066 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
LEUK_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.064483 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
TAOK2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.004673 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL54, O94957, Q6UW73, Q7LC09, Q9NSW2 | Gene names | TAOK2, KIAA0881, MAP3K17, PSK, PSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2) (Prostate-derived STE20-like kinase 1) (PSK-1) (Kinase from chicken homolog C) (hKFC-C). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.028448 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PATZ1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.002628 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBE1, Q9HBE2, Q9HBE3, Q9P1A9, Q9UDU0, Q9Y529 | Gene names | PATZ1, PATZ, RIAZ, ZBTB19, ZNF278, ZSG | |||
|
Domain Architecture |
|
|||||
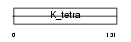
| Description | POZ-, AT hook-, and zinc finger-containing protein 1 (Zinc finger protein 278) (Zinc finger sarcoma gene protein) (BTB-POZ domain zinc finger transcription factor) (Protein kinase A RI-subunit alpha- associated protein) (Zinc finger and BTB domain-containing protein 19). | |||||
|
APBB2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.040644 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.012096 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.032791 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PCGF2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.038056 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |
|
|||||
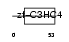
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.012022 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.032472 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
CSPG5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.055069 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.052620 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
CSPG5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.051905 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.018980 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
PCGF2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.034990 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |
|
|||||
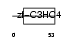
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.025515 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.033588 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.022798 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.048696 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.013119 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.024925 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.022613 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.009085 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
CE025_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.037677 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NDZ2, Q6NXN8, Q6ZTU4, Q8IZ15 | Gene names | C5orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf25. | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.015484 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.048057 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
LRCH4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.017823 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
PRR11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.031456 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HE9, Q9NUZ7, Q9NXE9 | Gene names | PRR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 11. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.038103 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SC24B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.021086 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95487 | Gene names | SEC24B | |||
|
Domain Architecture |
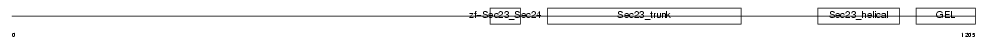
|
|||||
| Description | Protein transport protein Sec24B (SEC24-related protein B). | |||||
|
SUHW3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.005217 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P3Y5, Q6ZPM2, Q8K145 | Gene names | Suhw3, Kiaa1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
UBP2L_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.028833 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
ABLM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.007411 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.058327 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.044001 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CXX1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.044859 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15255 | Gene names | CXX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAAX box protein 1 (Cerebral protein 5). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.022821 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.021411 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
SON_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.040145 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TAF1C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.024753 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15572, Q59F67, Q8N6V3 | Gene names | TAF1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 110 kDa) (TAFI110). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.047224 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TNR21_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.015927 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.027401 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.012816 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.030948 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K1949_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.020286 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.012131 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PACS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.021367 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6VY07, Q6PJY6, Q6PKB6, Q7Z590, Q7Z5W4, Q8N8K6, Q96MW0, Q9NW92, Q9ULP5 | Gene names | PACS1, KIAA1175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
RNF12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.014825 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.016975 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
TAOK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.001710 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.035616 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.000366 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.017179 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MY116_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.026896 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P17564 | Gene names | Myd116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid differentiation primary response protein MyD116. | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.020761 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.020230 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.017509 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
SYRM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.014301 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T160, Q96FU5, Q9H8K8 | Gene names | RARSL, RARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable arginyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.19) (Arginine--tRNA ligase) (ArgRS) (Arginyl-tRNA synthetase-like). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.026659 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.028273 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TM131_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.021474 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.007207 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.014881 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.042463 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ANKS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.003937 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59672, Q6ZQG0 | Gene names | Anks1a, Anks1, Kiaa0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A. | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.009282 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
CPSF7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.014030 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
DEN1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.012772 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DIAP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.007907 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
EM55_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.004875 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70290 | Gene names | Mpp1 | |||
|
Domain Architecture |
|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1) (Palmitoylated protein p55). | |||||
|
HXA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.002334 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
MDM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.013513 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23804, Q61040, Q64330 | Gene names | Mdm2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.016665 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.014888 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.008224 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.010926 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.023437 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
SOS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.008123 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.021377 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ZN414_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.009462 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96IQ9 | Gene names | ZNF414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
ATS14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.003276 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.015055 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
CC054_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.013254 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96EL1 | Gene names | C3orf54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf54. | |||||
|
CN155_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.025341 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
K2027_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.019469 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.005416 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.018665 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.020924 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SGIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.014404 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
TB182_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.016936 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
TCF19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.012891 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KJ5, Q8BPA3, Q9CT93 | Gene names | Tcf19, Sc1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 19-like protein (SC1 protein homolog). | |||||
|
AP3M1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.193346 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2T2 | Gene names | AP3M1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.193214 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JKC8, Q5BKQ6 | Gene names | Ap3m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.191147 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53677 | Gene names | AP3M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B). | |||||
|
AP3M2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.192339 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R2R9, Q3UYJ3, Q923G7 | Gene names | Ap3m2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B) (m3B). | |||||
|
AP4M1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.264709 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JKC7 | Gene names | Ap4m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
STON2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
STON2_MOUSE
|
||||||
| NC score | 0.973763 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
STON1_HUMAN
|
||||||
| NC score | 0.906721 (rank : 3) | θ value | 2.70778e-113 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6Q2, Q96JE3, Q9BYX3 | Gene names | STON1, SALF, SBLF, STN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
STON1_MOUSE
|
||||||
| NC score | 0.904887 (rank : 4) | θ value | 1.58745e-113 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
AP2M1_HUMAN
|
||||||
| NC score | 0.469648 (rank : 5) | θ value | 1.09485e-13 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96CW1, P20172, P53679 | Gene names | AP2M1, CLAPM1, KIAA0109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (HA2 50 kDa subunit) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP2M1_MOUSE
|
||||||
| NC score | 0.469648 (rank : 6) | θ value | 1.09485e-13 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84091, P20172, P53679 | Gene names | Ap2m1, Clapm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP1M1_HUMAN
|
||||||
| NC score | 0.460841 (rank : 7) | θ value | 1.42992e-13 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BXS5 | Gene names | AP1M1, CLTNM | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M1_MOUSE
|
||||||
| NC score | 0.458140 (rank : 8) | θ value | 3.18553e-13 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35585 | Gene names | Ap1m1, Cltnm | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M2_HUMAN
|
||||||
| NC score | 0.448017 (rank : 9) | θ value | 5.43371e-13 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6Q5, Q9BSI8 | Gene names | AP1M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP1M2_MOUSE
|
||||||
| NC score | 0.447466 (rank : 10) | θ value | 9.26847e-13 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVP1, Q99LA4, Q9CWP7 | Gene names | Ap1m2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP4M1_HUMAN
|
||||||
| NC score | 0.304589 (rank : 11) | θ value | 0.0113563 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00189, Q9UHK9 | Gene names | AP4M1, MUARP2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP4M1_MOUSE
|
||||||
| NC score | 0.264709 (rank : 12) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JKC7 | Gene names | Ap4m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP3M1_HUMAN
|
||||||
| NC score | 0.193346 (rank : 13) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2T2 | Gene names | AP3M1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M1_MOUSE
|
||||||
| NC score | 0.193214 (rank : 14) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JKC8, Q5BKQ6 | Gene names | Ap3m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M2_MOUSE
|
||||||
| NC score | 0.192339 (rank : 15) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R2R9, Q3UYJ3, Q923G7 | Gene names | Ap3m2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B) (m3B). | |||||
|
AP3M2_HUMAN
|
||||||
| NC score | 0.191147 (rank : 16) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53677 | Gene names | AP3M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B). | |||||
|
PALB2_MOUSE
|
||||||
| NC score | 0.078085 (rank : 17) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.075374 (rank : 18) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.072474 (rank : 19) | θ value | 0.00665767 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.070160 (rank : 20) | θ value | 0.00228821 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
LEUK_MOUSE
|
||||||
| NC score | 0.064483 (rank : 21) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.064068 (rank : 22) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.061615 (rank : 23) | θ value | 0.00390308 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.058327 (rank : 24) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.055764 (rank : 25) | θ value | 0.00298849 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CSPG5_HUMAN
|
||||||
| NC score | 0.055069 (rank : 26) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.052620 (rank : 27) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
CSPG5_MOUSE
|
||||||
| NC score | 0.051905 (rank : 28) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.048696 (rank : 29) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.048057 (rank : 30) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.047224 (rank : 31) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CXX1_HUMAN
|
||||||
| NC score | 0.044859 (rank : 32) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15255 | Gene names | CXX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAAX box protein 1 (Cerebral protein 5). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.044001 (rank : 33) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.042463 (rank : 34) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.040644 (rank : 35) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
ECM2_HUMAN
|
||||||
| NC score | 0.040513 (rank : 36) | θ value | 0.000270298 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94769 | Gene names | ECM2 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 2 precursor (Matrix glycoprotein SC1/ECM2). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.040145 (rank : 37) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.039066 (rank : 38) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.038150 (rank : 39) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.038103 (rank : 40) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
PCGF2_HUMAN
|
||||||
| NC score | 0.038056 (rank : 41) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
CE025_HUMAN
|
||||||
| NC score | 0.037677 (rank : 42) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NDZ2, Q6NXN8, Q6ZTU4, Q8IZ15 | Gene names | C5orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf25. | |||||
|
EYA3_MOUSE
|
||||||
| NC score | 0.036151 (rank : 43) | θ value | 0.0330416 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.035616 (rank : 44) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
PCGF2_MOUSE
|
||||||
| NC score | 0.034990 (rank : 45) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.033588 (rank : 46) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.032791 (rank : 47) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.032472 (rank : 48) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
PRR11_HUMAN
|
||||||
| NC score | 0.031456 (rank : 49) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HE9, Q9NUZ7, Q9NXE9 | Gene names | PRR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 11. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.030948 (rank : 50) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
UBP2L_HUMAN
|
||||||
| NC score | 0.028833 (rank : 51) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.028448 (rank : 52) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.028273 (rank : 53) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.027401 (rank : 54) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
MY116_MOUSE
|
||||||
| NC score | 0.026896 (rank : 55) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P17564 | Gene names | Myd116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid differentiation primary response protein MyD116. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.026659 (rank : 56) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
BIRC2_HUMAN
|
||||||
| NC score | 0.026517 (rank : 57) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.025515 (rank : 58) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.025341 (rank : 59) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.024925 (rank : 60) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TAF1C_HUMAN
|
||||||
| NC score | 0.024753 (rank : 61) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15572, Q59F67, Q8N6V3 | Gene names | TAF1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 110 kDa) (TAFI110). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.023437 (rank : 62) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.022821 (rank : 63) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.022798 (rank : 64) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.022613 (rank : 65) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TM131_MOUSE
|
||||||
| NC score | 0.021474 (rank : 66) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.021411 (rank : 67) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.021377 (rank : 68) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
PACS1_HUMAN
|
||||||
| NC score | 0.021367 (rank : 69) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6VY07, Q6PJY6, Q6PKB6, Q7Z590, Q7Z5W4, Q8N8K6, Q96MW0, Q9NW92, Q9ULP5 | Gene names | PACS1, KIAA1175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
SC24B_HUMAN
|
||||||
| NC score | 0.021086 (rank : 70) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95487 | Gene names | SEC24B | |||
|
Domain Architecture |
|
|||||
| Description | Protein transport protein Sec24B (SEC24-related protein B). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.020924 (rank : 71) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.020761 (rank : 72) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
K1949_MOUSE
|
||||||
| NC score | 0.020286 (rank : 73) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.020230 (rank : 74) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.019469 (rank : 75) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.018980 (rank : 76) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.018665 (rank : 77) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
LRCH4_MOUSE
|
||||||
| NC score | 0.017823 (rank : 78) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.017509 (rank : 79) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.017179 (rank : 80) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.016975 (rank : 81) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.016936 (rank : 82) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.016665 (rank : 83) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
TNR21_MOUSE
|
||||||
| NC score | 0.015927 (rank : 84) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.015484 (rank : 85) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.015055 (rank : 86) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.014888 (rank : 87) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.014881 (rank : 88) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RNF12_MOUSE
|
||||||
| NC score | 0.014825 (rank : 89) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
SGIP1_HUMAN
|
||||||
| NC score | 0.014404 (rank : 90) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SYRM_HUMAN
|
||||||
| NC score | 0.014301 (rank : 91) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T160, Q96FU5, Q9H8K8 | Gene names | RARSL, RARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable arginyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.19) (Arginine--tRNA ligase) (ArgRS) (Arginyl-tRNA synthetase-like). | |||||
|
CPSF7_MOUSE
|
||||||
| NC score | 0.014030 (rank : 92) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
MDM2_MOUSE
|
||||||
| NC score | 0.013513 (rank : 93) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23804, Q61040, Q64330 | Gene names | Mdm2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein). | |||||
|
CC054_HUMAN
|
||||||
| NC score | 0.013254 (rank : 94) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96EL1 | Gene names | C3orf54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf54. | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.013119 (rank : 95) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
TCF19_MOUSE
|
||||||
| NC score | 0.012891 (rank : 96) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KJ5, Q8BPA3, Q9CT93 | Gene names | Tcf19, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19-like protein (SC1 protein homolog). | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.012816 (rank : 97) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
DEN1A_MOUSE
|
||||||
| NC score | 0.012772 (rank : 98) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.012131 (rank : 99) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NCA11_MOUSE
|
||||||
| NC score | 0.012096 (rank : 100) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.012022 (rank : 101) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.010926 (rank : 102) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
ZN414_HUMAN
|
||||||
| NC score | 0.009462 (rank : 103) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96IQ9 | Gene names | ZNF414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.009282 (rank : 104) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.009085 (rank : 105) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.008224 (rank : 106) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
SOS2_HUMAN
|
||||||
| NC score | 0.008123 (rank : 107) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
DIAP2_MOUSE
|
||||||
| NC score | 0.007907 (rank : 108) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
ABLM1_MOUSE
|
||||||
| NC score | 0.007411 (rank : 109) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.007207 (rank : 110) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.005416 (rank : 111) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
SUHW3_MOUSE
|
||||||
| NC score | 0.005217 (rank : 112) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P3Y5, Q6ZPM2, Q8K145 | Gene names | Suhw3, Kiaa1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
EM55_MOUSE
|
||||||
| NC score | 0.004875 (rank : 113) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70290 | Gene names | Mpp1 | |||
|
Domain Architecture |

|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1) (Palmitoylated protein p55). | |||||
|
TAOK2_HUMAN
|
||||||
| NC score | 0.004673 (rank : 114) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL54, O94957, Q6UW73, Q7LC09, Q9NSW2 | Gene names | TAOK2, KIAA0881, MAP3K17, PSK, PSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2) (Prostate-derived STE20-like kinase 1) (PSK-1) (Kinase from chicken homolog C) (hKFC-C). | |||||
|
ANKS1_MOUSE
|
||||||
| NC score | 0.003937 (rank : 115) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59672, Q6ZQG0 | Gene names | Anks1a, Anks1, Kiaa0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A. | |||||
|
ATS14_HUMAN
|
||||||
| NC score | 0.003276 (rank : 116) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
PATZ1_HUMAN
|
||||||
| NC score | 0.002628 (rank : 117) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBE1, Q9HBE2, Q9HBE3, Q9P1A9, Q9UDU0, Q9Y529 | Gene names | PATZ1, PATZ, RIAZ, ZBTB19, ZNF278, ZSG | |||
|
Domain Architecture |

|
|||||
| Description | POZ-, AT hook-, and zinc finger-containing protein 1 (Zinc finger protein 278) (Zinc finger sarcoma gene protein) (BTB-POZ domain zinc finger transcription factor) (Protein kinase A RI-subunit alpha- associated protein) (Zinc finger and BTB domain-containing protein 19). | |||||
|
HXA3_HUMAN
|
||||||
| NC score | 0.002334 (rank : 118) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
TAOK2_MOUSE
|
||||||
| NC score | 0.001710 (rank : 119) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.000366 (rank : 120) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||