Please be patient as the page loads
|
MAGD1_HUMAN
|
||||||
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MAGD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MAGD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.979967 (rank : 2) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 4.30305e-127 (rank : 3) | NC score | 0.951016 (rank : 7) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
TROP_HUMAN
|
||||||
| θ value | 6.25694e-102 (rank : 4) | NC score | 0.942214 (rank : 10) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
MAGD2_HUMAN
|
||||||
| θ value | 1.00126e-91 (rank : 5) | NC score | 0.975362 (rank : 3) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MAGL2_HUMAN
|
||||||
| θ value | 2.94719e-51 (rank : 6) | NC score | 0.950450 (rank : 8) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
MAGG1_HUMAN
|
||||||
| θ value | 9.48078e-50 (rank : 7) | NC score | 0.957255 (rank : 5) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96MG7, Q8IW16, Q8TEI6, Q9H214 | Gene names | NDNL2, HCA4, MAGEG1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2) (Hepatocellular carcinoma-associated protein 4). | |||||
|
MAGG1_MOUSE
|
||||||
| θ value | 8.02596e-49 (rank : 8) | NC score | 0.957781 (rank : 4) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 3.37202e-47 (rank : 9) | NC score | 0.674794 (rank : 36) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAGB3_HUMAN
|
||||||
| θ value | 1.12253e-42 (rank : 10) | NC score | 0.938689 (rank : 13) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
MAGBA_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 11) | NC score | 0.940510 (rank : 12) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96LZ2, Q494Y6, Q494Y7, Q9BZ78 | Gene names | MAGEB10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B10 (MAGE-B10 antigen). | |||||
|
MAGAB_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 12) | NC score | 0.919068 (rank : 25) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAGL2_MOUSE
|
||||||
| θ value | 1.3722e-40 (rank : 13) | NC score | 0.940962 (rank : 11) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QZ04 | Gene names | Magel2 | |||
|
Domain Architecture |
|
|||||
| Description | MAGE-like protein 2 (Protein nS7). | |||||
|
MAGB4_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 14) | NC score | 0.935602 (rank : 16) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAGBI_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 15) | NC score | 0.932749 (rank : 18) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
MAGF1_HUMAN
|
||||||
| θ value | 1.6774e-38 (rank : 16) | NC score | 0.956461 (rank : 6) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAY2, Q9H215 | Gene names | MAGEF1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen F1 (MAGE-F1 antigen). | |||||
|
MAGB1_HUMAN
|
||||||
| θ value | 2.42216e-37 (rank : 17) | NC score | 0.931945 (rank : 19) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
MAGA1_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 18) | NC score | 0.907780 (rank : 27) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43355, O00346 | Gene names | MAGEA1, MAGE1, MAGE1A | |||
|
Domain Architecture |
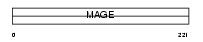
|
|||||
| Description | Melanoma-associated antigen 1 (MAGE-1 antigen) (Antigen MZ2-E). | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 19) | NC score | 0.925420 (rank : 22) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MAGB2_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 20) | NC score | 0.935662 (rank : 15) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |
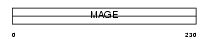
|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
MAGAA_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 21) | NC score | 0.919618 (rank : 24) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |
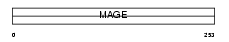
|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
NECD_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 22) | NC score | 0.942436 (rank : 9) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |
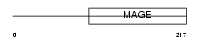
|
|||||
| Description | Necdin. | |||||
|
MAGA4_HUMAN
|
||||||
| θ value | 3.86705e-35 (rank : 23) | NC score | 0.903067 (rank : 28) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43358, Q14798 | Gene names | MAGEA4, MAGE4 | |||
|
Domain Architecture |
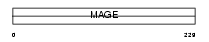
|
|||||
| Description | Melanoma-associated antigen 4 (MAGE-4 antigen) (MAGE-X2) (MAGE-41). | |||||
|
MAGE2_HUMAN
|
||||||
| θ value | 8.61488e-35 (rank : 24) | NC score | 0.934723 (rank : 17) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TD90 | Gene names | MAGEE2, HCA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E2 (MAGE-E2 antigen) (Hepatocellular carcinoma-associated protein 3). | |||||
|
NECD_MOUSE
|
||||||
| θ value | 7.29293e-34 (rank : 25) | NC score | 0.937461 (rank : 14) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 26) | NC score | 0.872509 (rank : 33) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAGA6_HUMAN
|
||||||
| θ value | 7.54701e-31 (rank : 27) | NC score | 0.892110 (rank : 29) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43360 | Gene names | MAGEA6, MAGE6 | |||
|
Domain Architecture |
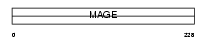
|
|||||
| Description | Melanoma-associated antigen 6 (MAGE-6 antigen) (MAGE3B). | |||||
|
MAGAC_HUMAN
|
||||||
| θ value | 7.54701e-31 (rank : 28) | NC score | 0.891272 (rank : 30) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |
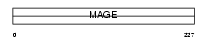
|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
MAGA9_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 29) | NC score | 0.911476 (rank : 26) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43362, Q92910 | Gene names | MAGEA9, MAGE9 | |||
|
Domain Architecture |
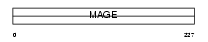
|
|||||
| Description | Melanoma-associated antigen 9 (MAGE-9 antigen). | |||||
|
MAGC2_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 30) | NC score | 0.924689 (rank : 23) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UBF1, Q5JZ00, Q96D45, Q9P1M6, Q9P1M7 | Gene names | MAGEC2, HCA587, MAGEE1 | |||
|
Domain Architecture |
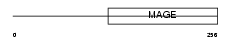
|
|||||
| Description | Melanoma-associated antigen C2 (MAGE-C2 antigen) (MAGE-E1 antigen) (Hepatocellular carcinoma-associated antigen 587) (Cancer-testis antigen 10) (CT10). | |||||
|
MAGA3_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 31) | NC score | 0.889378 (rank : 31) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43357 | Gene names | MAGEA3, MAGE3 | |||
|
Domain Architecture |
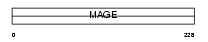
|
|||||
| Description | Melanoma-associated antigen 3 (MAGE-3 antigen) (Antigen MZ2-D). | |||||
|
MAGH1_MOUSE
|
||||||
| θ value | 1.92365e-26 (rank : 32) | NC score | 0.928162 (rank : 21) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NWG9, Q99PB0 | Gene names | Mageh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen). | |||||
|
MAGH1_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 33) | NC score | 0.928717 (rank : 20) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H213, Q5JRJ3, Q9Y5M2 | Gene names | MAGEH1, APR1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen) (Restin) (Apoptosis- related protein 1) (APR-1). | |||||
|
MAGA2_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 34) | NC score | 0.888061 (rank : 32) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43356 | Gene names | MAGEA2, MAGE2, MAGEA2A | |||
|
Domain Architecture |
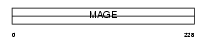
|
|||||
| Description | Melanoma-associated antigen 2 (MAGE-2 antigen). | |||||
|
MAGC3_HUMAN
|
||||||
| θ value | 1.74391e-19 (rank : 35) | NC score | 0.858983 (rank : 34) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
MAGA8_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 36) | NC score | 0.858129 (rank : 35) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |
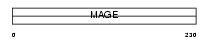
|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 37) | NC score | 0.060908 (rank : 48) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 38) | NC score | 0.016451 (rank : 115) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 39) | NC score | 0.096590 (rank : 38) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 40) | NC score | 0.053415 (rank : 56) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 41) | NC score | 0.037306 (rank : 80) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 42) | NC score | 0.023443 (rank : 94) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 43) | NC score | 0.041797 (rank : 72) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
EMIL2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 44) | NC score | 0.016918 (rank : 111) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 45) | NC score | 0.020382 (rank : 103) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 46) | NC score | 0.061038 (rank : 46) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 47) | NC score | 0.058939 (rank : 52) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 48) | NC score | 0.004501 (rank : 138) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.041405 (rank : 73) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
NMDE3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.013574 (rank : 122) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
TM16H_HUMAN
|
||||||
| θ value | 0.163984 (rank : 51) | NC score | 0.035586 (rank : 82) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 52) | NC score | 0.020840 (rank : 102) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 0.21417 (rank : 53) | NC score | 0.043133 (rank : 66) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 54) | NC score | 0.050052 (rank : 60) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.043003 (rank : 68) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.031849 (rank : 85) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.038106 (rank : 79) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.365318 (rank : 58) | NC score | 0.042194 (rank : 71) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
ALEX_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.039832 (rank : 75) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
CASL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.026578 (rank : 91) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |
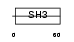
|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.017575 (rank : 110) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.004314 (rank : 139) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.018451 (rank : 107) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
DTX1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.023383 (rank : 95) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |
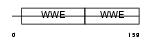
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.039126 (rank : 77) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.022091 (rank : 100) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.017787 (rank : 109) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.040666 (rank : 74) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
DREB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.022354 (rank : 99) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
DREB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.021951 (rank : 101) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
LPHN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.006947 (rank : 132) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.030274 (rank : 88) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SELV_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.043678 (rank : 64) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
SIA7A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.012305 (rank : 124) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.050478 (rank : 59) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.031213 (rank : 87) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.008553 (rank : 131) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.014281 (rank : 121) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.011384 (rank : 126) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
MEF2B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.023283 (rank : 97) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
TSYL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.018205 (rank : 108) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88852, Q3TKW0 | Gene names | Tspyl1, Tspyl | |||
|
Domain Architecture |
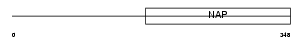
|
|||||
| Description | Testis-specific Y-encoded-like protein 1 (TSPY-like 1). | |||||
|
TX13A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.016472 (rank : 114) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXU3, Q32NB6 | Gene names | TEX13A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 13A protein. | |||||
|
UBP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.004561 (rank : 137) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
ASAH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.011801 (rank : 125) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NR71, Q3KNU1, Q5SNT7, Q5SZP6, Q5SZP7, Q5T1D5, Q71ME6 | Gene names | ASAH2, HNAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neutral ceramidase (EC 3.5.1.23) (NCDase) (N-CDase) (Acylsphingosine deacylase 2) (N-acylsphingosine amidohydrolase 2) (BCDase) (LCDase) (hCD) [Contains: Neutral ceramidase soluble form]. | |||||
|
CHCH2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.018509 (rank : 106) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D1L0, Q3TUG8 | Gene names | Chchd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 2. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.049502 (rank : 61) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.014838 (rank : 119) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CXA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.004931 (rank : 136) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6H8, Q9H537 | Gene names | GJA3 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-3 protein (Connexin-46) (Cx46). | |||||
|
NUFP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.015933 (rank : 117) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z417, Q9P2M5 | Gene names | NUFIP2, KIAA1321, PIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 2 (FMRP- interacting protein 2) (82 kDa FMRP-interacting protein) (82-FIP) (Proliferation-inducing gene 1 protein). | |||||
|
OMGP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.001737 (rank : 142) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q63912 | Gene names | Omg, Omgp | |||
|
Domain Architecture |
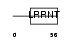
|
|||||
| Description | Oligodendrocyte-myelin glycoprotein precursor. | |||||
|
SF01_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.043000 (rank : 69) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SF01_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.035386 (rank : 83) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
TICN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.006336 (rank : 133) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |
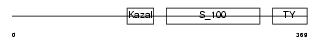
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.016027 (rank : 116) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.001777 (rank : 141) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.015800 (rank : 118) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.020238 (rank : 104) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.045634 (rank : 62) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.024147 (rank : 93) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.028443 (rank : 89) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.024344 (rank : 92) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
IF35_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.043280 (rank : 65) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.012958 (rank : 123) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.016859 (rank : 112) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.039691 (rank : 76) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.014299 (rank : 120) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.038613 (rank : 78) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
WDR33_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.010495 (rank : 127) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
BCAR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.022918 (rank : 98) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.036407 (rank : 81) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DAB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.016701 (rank : 113) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |
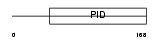
|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.043088 (rank : 67) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
HIG2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.009423 (rank : 130) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQJ1 | Gene names | Higd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIG1 domain family member 2A. | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.010440 (rank : 129) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.031778 (rank : 86) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.027527 (rank : 90) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.032543 (rank : 84) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.019556 (rank : 105) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.004201 (rank : 140) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.043819 (rank : 63) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
ZN672_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | -0.005787 (rank : 147) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q499Z4, Q96H65, Q96IM3, Q9H6G5 | Gene names | ZNF672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 672. | |||||
|
AAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.005172 (rank : 134) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
BAI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.005078 (rank : 135) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
PI51C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.010494 (rank : 128) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | -0.000860 (rank : 144) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
RIPK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | -0.004197 (rank : 146) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y572 | Gene names | RIPK3, RIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.023321 (rank : 96) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.042542 (rank : 70) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TENA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | -0.000911 (rank : 145) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
WDHD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.001248 (rank : 143) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.085198 (rank : 40) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052991 (rank : 57) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051900 (rank : 58) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
LORI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.094015 (rank : 39) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LORI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.057238 (rank : 53) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
MAGA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.474606 (rank : 37) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P43359 | Gene names | MAGEA5, MAGE5 | |||
|
Domain Architecture |
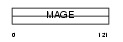
|
|||||
| Description | Melanoma-associated antigen 5 (MAGE-5 antigen). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.065278 (rank : 42) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.062044 (rank : 45) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.060771 (rank : 49) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.055178 (rank : 54) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NU214_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.064685 (rank : 44) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PHXR5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.074353 (rank : 41) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.060986 (rank : 47) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PO121_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.059959 (rank : 51) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
SELPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.065243 (rank : 43) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.054087 (rank : 55) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.060349 (rank : 50) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MAGD1_MOUSE
|
||||||
| NC score | 0.979967 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
MAGD2_HUMAN
|
||||||
| NC score | 0.975362 (rank : 3) | θ value | 1.00126e-91 (rank : 5) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MAGG1_MOUSE
|
||||||
| NC score | 0.957781 (rank : 4) | θ value | 8.02596e-49 (rank : 8) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
MAGG1_HUMAN
|
||||||
| NC score | 0.957255 (rank : 5) | θ value | 9.48078e-50 (rank : 7) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96MG7, Q8IW16, Q8TEI6, Q9H214 | Gene names | NDNL2, HCA4, MAGEG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2) (Hepatocellular carcinoma-associated protein 4). | |||||
|
MAGF1_HUMAN
|
||||||
| NC score | 0.956461 (rank : 6) | θ value | 1.6774e-38 (rank : 16) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAY2, Q9H215 | Gene names | MAGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen F1 (MAGE-F1 antigen). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.951016 (rank : 7) | θ value | 4.30305e-127 (rank : 3) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
MAGL2_HUMAN
|
||||||
| NC score | 0.950450 (rank : 8) | θ value | 2.94719e-51 (rank : 6) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
NECD_HUMAN
|
||||||
| NC score | 0.942436 (rank : 9) | θ value | 2.96089e-35 (rank : 22) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
TROP_HUMAN
|
||||||
| NC score | 0.942214 (rank : 10) | θ value | 6.25694e-102 (rank : 4) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
MAGL2_MOUSE
|
||||||
| NC score | 0.940962 (rank : 11) | θ value | 1.3722e-40 (rank : 13) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QZ04 | Gene names | Magel2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Protein nS7). | |||||
|
MAGBA_HUMAN
|
||||||
| NC score | 0.940510 (rank : 12) | θ value | 2.11701e-41 (rank : 11) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96LZ2, Q494Y6, Q494Y7, Q9BZ78 | Gene names | MAGEB10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B10 (MAGE-B10 antigen). | |||||
|
MAGB3_HUMAN
|
||||||
| NC score | 0.938689 (rank : 13) | θ value | 1.12253e-42 (rank : 10) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
NECD_MOUSE
|
||||||
| NC score | 0.937461 (rank : 14) | θ value | 7.29293e-34 (rank : 25) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MAGB2_HUMAN
|
||||||
| NC score | 0.935662 (rank : 15) | θ value | 1.32909e-35 (rank : 20) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
MAGB4_HUMAN
|
||||||
| NC score | 0.935602 (rank : 16) | θ value | 2.34062e-40 (rank : 14) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAGE2_HUMAN
|
||||||
| NC score | 0.934723 (rank : 17) | θ value | 8.61488e-35 (rank : 24) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TD90 | Gene names | MAGEE2, HCA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E2 (MAGE-E2 antigen) (Hepatocellular carcinoma-associated protein 3). | |||||
|
MAGBI_HUMAN
|
||||||
| NC score | 0.932749 (rank : 18) | θ value | 1.98146e-39 (rank : 15) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
MAGB1_HUMAN
|
||||||
| NC score | 0.931945 (rank : 19) | θ value | 2.42216e-37 (rank : 17) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
MAGH1_HUMAN
|
||||||
| NC score | 0.928717 (rank : 20) | θ value | 5.59698e-26 (rank : 33) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H213, Q5JRJ3, Q9Y5M2 | Gene names | MAGEH1, APR1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen) (Restin) (Apoptosis- related protein 1) (APR-1). | |||||
|
MAGH1_MOUSE
|
||||||
| NC score | 0.928162 (rank : 21) | θ value | 1.92365e-26 (rank : 32) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NWG9, Q99PB0 | Gene names | Mageh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen). | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.925420 (rank : 22) | θ value | 5.96599e-36 (rank : 19) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MAGC2_HUMAN
|
||||||
| NC score | 0.924689 (rank : 23) | θ value | 1.08979e-29 (rank : 30) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UBF1, Q5JZ00, Q96D45, Q9P1M6, Q9P1M7 | Gene names | MAGEC2, HCA587, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C2 (MAGE-C2 antigen) (MAGE-E1 antigen) (Hepatocellular carcinoma-associated antigen 587) (Cancer-testis antigen 10) (CT10). | |||||
|
MAGAA_HUMAN
|
||||||
| NC score | 0.919618 (rank : 24) | θ value | 2.96089e-35 (rank : 21) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
MAGAB_HUMAN
|
||||||
| NC score | 0.919068 (rank : 25) | θ value | 4.71619e-41 (rank : 12) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAGA9_HUMAN
|
||||||
| NC score | 0.911476 (rank : 26) | θ value | 2.86786e-30 (rank : 29) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43362, Q92910 | Gene names | MAGEA9, MAGE9 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 9 (MAGE-9 antigen). | |||||
|
MAGA1_HUMAN
|
||||||
| NC score | 0.907780 (rank : 27) | θ value | 5.96599e-36 (rank : 18) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43355, O00346 | Gene names | MAGEA1, MAGE1, MAGE1A | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 1 (MAGE-1 antigen) (Antigen MZ2-E). | |||||
|
MAGA4_HUMAN
|
||||||
| NC score | 0.903067 (rank : 28) | θ value | 3.86705e-35 (rank : 23) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43358, Q14798 | Gene names | MAGEA4, MAGE4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 4 (MAGE-4 antigen) (MAGE-X2) (MAGE-41). | |||||
|
MAGA6_HUMAN
|
||||||
| NC score | 0.892110 (rank : 29) | θ value | 7.54701e-31 (rank : 27) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43360 | Gene names | MAGEA6, MAGE6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 6 (MAGE-6 antigen) (MAGE3B). | |||||
|
MAGAC_HUMAN
|
||||||
| NC score | 0.891272 (rank : 30) | θ value | 7.54701e-31 (rank : 28) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
MAGA3_HUMAN
|
||||||
| NC score | 0.889378 (rank : 31) | θ value | 5.06226e-27 (rank : 31) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P43357 | Gene names | MAGEA3, MAGE3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 3 (MAGE-3 antigen) (Antigen MZ2-D). | |||||
|
MAGA2_HUMAN
|
||||||
| NC score | 0.888061 (rank : 32) | θ value | 2.12685e-25 (rank : 34) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43356 | Gene names | MAGEA2, MAGE2, MAGEA2A | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 2 (MAGE-2 antigen). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.872509 (rank : 33) | θ value | 5.22648e-32 (rank : 26) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAGC3_HUMAN
|
||||||
| NC score | 0.858983 (rank : 34) | θ value | 1.74391e-19 (rank : 35) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
MAGA8_HUMAN
|
||||||
| NC score | 0.858129 (rank : 35) | θ value | 4.02038e-16 (rank : 36) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.674794 (rank : 36) | θ value | 3.37202e-47 (rank : 9) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAGA5_HUMAN
|
||||||
| NC score | 0.474606 (rank : 37) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P43359 | Gene names | MAGEA5, MAGE5 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 5 (MAGE-5 antigen). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.096590 (rank : 38) | θ value | 0.00665767 (rank : 39) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.094015 (rank : 39) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.085198 (rank : 40) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PHXR5_MOUSE
|
||||||
| NC score | 0.074353 (rank : 41) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.065278 (rank : 42) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.065243 (rank : 43) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.064685 (rank : 44) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.062044 (rank : 45) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.061038 (rank : 46) | θ value | 0.0961366 (rank : 46) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.060986 (rank : 47) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.060908 (rank : 48) | θ value | 0.00035302 (rank : 37) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.060771 (rank : 49) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.060349 (rank : 50) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.059959 (rank : 51) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.058939 (rank : 52) | θ value | 0.125558 (rank : 47) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.057238 (rank : 53) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.055178 (rank : 54) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.054087 (rank : 55) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.053415 (rank : 56) | θ value | 0.0148317 (rank : 40) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.052991 (rank : 57) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.051900 (rank : 58) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.050478 (rank : 59) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.050052 (rank : 60) | θ value | 0.21417 (rank : 54) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.049502 (rank : 61) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.045634 (rank : 62) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.043819 (rank : 63) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.043678 (rank : 64) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.043280 (rank : 65) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.043133 (rank : 66) | θ value | 0.21417 (rank : 53) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.043088 (rank : 67) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.043003 (rank : 68) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.043000 (rank : 69) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.042542 (rank : 70) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.042194 (rank : 71) | θ value | 0.365318 (rank : 58) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.041797 (rank : 72) | θ value | 0.0563607 (rank : 43) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.041405 (rank : 73) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.040666 (rank : 74) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.039832 (rank : 75) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.039691 (rank : 76) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.039126 (rank : 77) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.038613 (rank : 78) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.038106 (rank : 79) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.037306 (rank : 80) | θ value | 0.0193708 (rank : 41) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.036407 (rank : 81) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
TM16H_HUMAN
|
||||||
| NC score | 0.035586 (rank : 82) | θ value | 0.163984 (rank : 51) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
SF01_MOUSE
|
||||||
| NC score | 0.035386 (rank : 83) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.032543 (rank : 84) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.031849 (rank : 85) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.031778 (rank : 86) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.031213 (rank : 87) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.030274 (rank : 88) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.028443 (rank : 89) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.027527 (rank : 90) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
CASL_HUMAN
|
||||||
| NC score | 0.026578 (rank : 91) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.024344 (rank : 92) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.024147 (rank : 93) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.023443 (rank : 94) | θ value | 0.0431538 (rank : 42) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
DTX1_MOUSE
|
||||||
| NC score | 0.023383 (rank : 95) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.023321 (rank : 96) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MEF2B_HUMAN
|
||||||
| NC score | 0.023283 (rank : 97) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
BCAR1_MOUSE
|
||||||
| NC score | 0.022918 (rank : 98) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
DREB_HUMAN
|
||||||
| NC score | 0.022354 (rank : 99) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.022091 (rank : 100) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
DREB_MOUSE
|
||||||
| NC score | 0.021951 (rank : 101) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.020840 (rank : 102) | θ value | 0.21417 (rank : 52) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.020382 (rank : 103) | θ value | 0.0961366 (rank : 45) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.020238 (rank : 104) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.019556 (rank : 105) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
CHCH2_MOUSE
|
||||||
| NC score | 0.018509 (rank : 106) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D1L0, Q3TUG8 | Gene names | Chchd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 2. | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.018451 (rank : 107) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
TSYL1_MOUSE
|
||||||
| NC score | 0.018205 (rank : 108) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88852, Q3TKW0 | Gene names | Tspyl1, Tspyl | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific Y-encoded-like protein 1 (TSPY-like 1). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.017787 (rank : 109) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.017575 (rank : 110) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
EMIL2_MOUSE
|
||||||
| NC score | 0.016918 (rank : 111) | θ value | 0.0961366 (rank : 44) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.016859 (rank : 112) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
DAB2_MOUSE
|
||||||
| NC score | 0.016701 (rank : 113) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
TX13A_HUMAN
|
||||||
| NC score | 0.016472 (rank : 114) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXU3, Q32NB6 | Gene names | TEX13A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 13A protein. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.016451 (rank : 115) | θ value | 0.00390308 (rank : 38) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.016027 (rank : 116) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
NUFP2_HUMAN
|
||||||
| NC score | 0.015933 (rank : 117) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z417, Q9P2M5 | Gene names | NUFIP2, KIAA1321, PIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 2 (FMRP- interacting protein 2) (82 kDa FMRP-interacting protein) (82-FIP) (Proliferation-inducing gene 1 protein). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.015800 (rank : 118) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.014838 (rank : 119) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.014299 (rank : 120) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
NCA11_MOUSE
|
||||||
| NC score | 0.014281 (rank : 121) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
NMDE3_MOUSE
|
||||||
| NC score | 0.013574 (rank : 122) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.012958 (rank : 123) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
SIA7A_HUMAN
|
||||||
| NC score | 0.012305 (rank : 124) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
ASAH2_HUMAN
|
||||||
| NC score | 0.011801 (rank : 125) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NR71, Q3KNU1, Q5SNT7, Q5SZP6, Q5SZP7, Q5T1D5, Q71ME6 | Gene names | ASAH2, HNAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neutral ceramidase (EC 3.5.1.23) (NCDase) (N-CDase) (Acylsphingosine deacylase 2) (N-acylsphingosine amidohydrolase 2) (BCDase) (LCDase) (hCD) [Contains: Neutral ceramidase soluble form]. | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.011384 (rank : 126) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
WDR33_HUMAN
|
||||||
| NC score | 0.010495 (rank : 127) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
PI51C_HUMAN
|
||||||
| NC score | 0.010494 (rank : 128) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.010440 (rank : 129) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
HIG2A_MOUSE
|
||||||
| NC score | 0.009423 (rank : 130) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQJ1 | Gene names | Higd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIG1 domain family member 2A. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.008553 (rank : 131) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
LPHN1_HUMAN
|
||||||
| NC score | 0.006947 (rank : 132) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
TICN2_MOUSE
|
||||||
| NC score | 0.006336 (rank : 133) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
AAK1_HUMAN
|
||||||
| NC score | 0.005172 (rank : 134) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.005078 (rank : 135) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
CXA3_HUMAN
|
||||||
| NC score | 0.004931 (rank : 136) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6H8, Q9H537 | Gene names | GJA3 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-3 protein (Connexin-46) (Cx46). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.004561 (rank : 137) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.004501 (rank : 138) | θ value | 0.163984 (rank : 48) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.004314 (rank : 139) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.004201 (rank : 140) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.001777 (rank : 141) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
OMGP_MOUSE
|
||||||
| NC score | 0.001737 (rank : 142) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q63912 | Gene names | Omg, Omgp | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte-myelin glycoprotein precursor. | |||||
|
WDHD1_MOUSE
|
||||||
| NC score | 0.001248 (rank : 143) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | -0.000860 (rank : 144) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
TENA_MOUSE
|
||||||
| NC score | -0.000911 (rank : 145) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
RIPK3_HUMAN
|
||||||
| NC score | -0.004197 (rank : 146) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y572 | Gene names | RIPK3, RIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3). | |||||
|
ZN672_HUMAN
|
||||||
| NC score | -0.005787 (rank : 147) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q499Z4, Q96H65, Q96IM3, Q9H6G5 | Gene names | ZNF672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 672. | |||||