Please be patient as the page loads
|
NMDE3_MOUSE
|
||||||
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NMDE1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.986784 (rank : 7) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMDE1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987585 (rank : 4) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.987393 (rank : 5) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.987198 (rank : 6) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.995986 (rank : 2) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.989017 (rank : 3) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE4_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.986577 (rank : 8) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMDZ1_MOUSE
|
||||||
| θ value | 2.31901e-72 (rank : 9) | NC score | 0.910246 (rank : 13) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_HUMAN
|
||||||
| θ value | 3.95564e-72 (rank : 10) | NC score | 0.910673 (rank : 12) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMD3A_HUMAN
|
||||||
| θ value | 2.56397e-71 (rank : 11) | NC score | 0.934229 (rank : 9) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMD3B_HUMAN
|
||||||
| θ value | 4.09345e-69 (rank : 12) | NC score | 0.926650 (rank : 11) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
NMD3B_MOUSE
|
||||||
| θ value | 5.91091e-68 (rank : 13) | NC score | 0.927284 (rank : 10) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
GRIA1_MOUSE
|
||||||
| θ value | 1.788e-48 (rank : 14) | NC score | 0.837557 (rank : 24) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_HUMAN
|
||||||
| θ value | 4.40402e-47 (rank : 15) | NC score | 0.836104 (rank : 26) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA4_HUMAN
|
||||||
| θ value | 1.28137e-46 (rank : 16) | NC score | 0.834166 (rank : 27) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA4_MOUSE
|
||||||
| θ value | 2.18568e-46 (rank : 17) | NC score | 0.832091 (rank : 28) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIK1_HUMAN
|
||||||
| θ value | 5.95217e-44 (rank : 18) | NC score | 0.841540 (rank : 18) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIK4_MOUSE
|
||||||
| θ value | 7.77379e-44 (rank : 19) | NC score | 0.841366 (rank : 19) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIK4_HUMAN
|
||||||
| θ value | 2.26182e-43 (rank : 20) | NC score | 0.841598 (rank : 17) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIA2_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 21) | NC score | 0.831727 (rank : 29) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA2_MOUSE
|
||||||
| θ value | 8.59492e-43 (rank : 22) | NC score | 0.831056 (rank : 30) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIK2_HUMAN
|
||||||
| θ value | 1.46607e-42 (rank : 23) | NC score | 0.840176 (rank : 20) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIK3_HUMAN
|
||||||
| θ value | 9.50281e-42 (rank : 24) | NC score | 0.842030 (rank : 15) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRIA3_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 25) | NC score | 0.830906 (rank : 31) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIK5_HUMAN
|
||||||
| θ value | 6.15952e-41 (rank : 26) | NC score | 0.840139 (rank : 21) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| θ value | 8.04462e-41 (rank : 27) | NC score | 0.839710 (rank : 22) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIA3_MOUSE
|
||||||
| θ value | 3.99248e-40 (rank : 28) | NC score | 0.830896 (rank : 32) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRID2_MOUSE
|
||||||
| θ value | 2.42216e-37 (rank : 29) | NC score | 0.843151 (rank : 14) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID2_HUMAN
|
||||||
| θ value | 3.16345e-37 (rank : 30) | NC score | 0.841981 (rank : 16) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID1_MOUSE
|
||||||
| θ value | 3.86705e-35 (rank : 31) | NC score | 0.839067 (rank : 23) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRID1_HUMAN
|
||||||
| θ value | 2.12192e-33 (rank : 32) | NC score | 0.837227 (rank : 25) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK1_MOUSE
|
||||||
| θ value | 2.34606e-32 (rank : 33) | NC score | 0.823029 (rank : 33) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
TS1R1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 34) | NC score | 0.062096 (rank : 35) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
TS1R1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 35) | NC score | 0.042524 (rank : 38) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 36) | NC score | 0.021588 (rank : 45) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 37) | NC score | 0.022614 (rank : 42) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
EFS_MOUSE
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.021954 (rank : 44) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
ERB1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.136239 (rank : 34) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
MAGD1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.013574 (rank : 58) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
RSPO4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.015800 (rank : 51) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJ73 | Gene names | Rspo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (Cysteine-rich and single thrombospondin domain-containing protein 4) (Cristin-4) (mCristin-4). | |||||
|
LAMC3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.004142 (rank : 75) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.012254 (rank : 60) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SHAN2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.015526 (rank : 53) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.010021 (rank : 63) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
TB182_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.018380 (rank : 46) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.016198 (rank : 50) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.014502 (rank : 54) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
KCC2G_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | -0.003310 (rank : 86) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13555, O15378, Q13556, Q5SQZ4 | Gene names | CAMK2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II gamma chain (EC 2.7.11.17) (CaM-kinase II gamma chain) (CaM kinase II gamma subunit) (CaMK-II subunit gamma). | |||||
|
TS13_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.029590 (rank : 40) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
CDT1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.017165 (rank : 48) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
CK056_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.018054 (rank : 47) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N612, Q9C0A4, Q9H0N3, Q9H624 | Gene names | C11orf56, KIAA1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.005922 (rank : 73) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CHSP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.023758 (rank : 41) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CR86 | Gene names | Carhsp1 | |||
|
Domain Architecture |
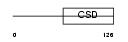
|
|||||
| Description | Calcium-regulated heat stable protein 1 (Calcium-regulated heat-stable protein of 24 kDa) (CRHSP-24). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.009804 (rank : 64) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.014317 (rank : 56) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
VINEX_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.006544 (rank : 69) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | -0.003357 (rank : 87) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
SHAN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.014439 (rank : 55) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.013194 (rank : 59) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | -0.002026 (rank : 84) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.006413 (rank : 70) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TSKS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.007842 (rank : 67) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJT2, Q8WXJ0 | Gene names | STK22S1, TSKS, TSKS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.006362 (rank : 72) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.017108 (rank : 49) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ENW1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.008696 (rank : 65) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQF0, O95244, O95245, Q8NHY7, Q9NRZ2, Q9NZG3 | Gene names | ERVWE1 | |||
|
Domain Architecture |
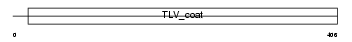
|
|||||
| Description | HERV-W_7q21.2 provirus ancestral Env polyprotein precursor (Envelope polyprotein gPr73) (HERV-7q Envelope protein) (HERV-W envelope protein) (Syncytin) (Syncytin-1) (Enverin) (Env-W) [Contains: Surface protein (SU) (gp50); Transmembrane protein (TM) (gp24)]. | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.022106 (rank : 43) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.010545 (rank : 62) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TAF6L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.007226 (rank : 68) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |
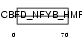
|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
TENS4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.006397 (rank : 71) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
ZEP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | -0.002067 (rank : 85) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
CK056_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.013820 (rank : 57) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U2I3, Q3TWF5, Q6P543, Q80T93, Q811J6, Q8BI34, Q8VE55 | Gene names | Kiaa1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56 homolog. | |||||
|
DLEC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.036094 (rank : 39) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLA1 | Gene names | Dlec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in lung and esophageal cancer protein 1 homolog. | |||||
|
NAB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.005625 (rank : 74) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15742, O76006, Q14797 | Gene names | NAB2, MADER | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2) (Melanoma- associated delayed early response protein) (Protein MADER). | |||||
|
NRPN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | -0.001218 (rank : 83) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61955 | Gene names | Klk8, Nrpn, Prss19 | |||
|
Domain Architecture |
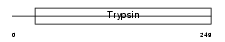
|
|||||
| Description | Neuropsin precursor (EC 3.4.21.-) (NP) (Kallikrein-8). | |||||
|
SON_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.011198 (rank : 61) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ADA25_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.000006 (rank : 80) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R159, Q9D4E4 | Gene names | Adam25 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 25 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 25) (Testase 2). | |||||
|
BTBD7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.003562 (rank : 76) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P203, Q69Z05, Q7Z308, Q86TS0, Q9HAA4, Q9NVM0 | Gene names | BTBD7, KIAA1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
CIAS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.002786 (rank : 77) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
DLX5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | -0.000745 (rank : 81) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56178, Q9UPL1 | Gene names | DLX5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
DLX5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | -0.000805 (rank : 82) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
EFS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.015722 (rank : 52) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.008596 (rank : 66) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
MMP11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.000759 (rank : 79) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | -0.004104 (rank : 88) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.002689 (rank : 78) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
GABR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.053306 (rank : 36) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053173 (rank : 37) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
NMDE3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.995986 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.989017 (rank : 3) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE1_MOUSE
|
||||||
| NC score | 0.987585 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| NC score | 0.987393 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| NC score | 0.987198 (rank : 6) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE1_HUMAN
|
||||||
| NC score | 0.986784 (rank : 7) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMDE4_MOUSE
|
||||||
| NC score | 0.986577 (rank : 8) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMD3A_HUMAN
|
||||||
| NC score | 0.934229 (rank : 9) | θ value | 2.56397e-71 (rank : 11) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMD3B_MOUSE
|
||||||
| NC score | 0.927284 (rank : 10) | θ value | 5.91091e-68 (rank : 13) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| NC score | 0.926650 (rank : 11) | θ value | 4.09345e-69 (rank : 12) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
NMDZ1_HUMAN
|
||||||
| NC score | 0.910673 (rank : 12) | θ value | 3.95564e-72 (rank : 10) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| NC score | 0.910246 (rank : 13) | θ value | 2.31901e-72 (rank : 9) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
GRID2_MOUSE
|
||||||
| NC score | 0.843151 (rank : 14) | θ value | 2.42216e-37 (rank : 29) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIK3_HUMAN
|
||||||
| NC score | 0.842030 (rank : 15) | θ value | 9.50281e-42 (rank : 24) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRID2_HUMAN
|
||||||
| NC score | 0.841981 (rank : 16) | θ value | 3.16345e-37 (rank : 30) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIK4_HUMAN
|
||||||
| NC score | 0.841598 (rank : 17) | θ value | 2.26182e-43 (rank : 20) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK1_HUMAN
|
||||||
| NC score | 0.841540 (rank : 18) | θ value | 5.95217e-44 (rank : 18) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIK4_MOUSE
|
||||||
| NC score | 0.841366 (rank : 19) | θ value | 7.77379e-44 (rank : 19) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIK2_HUMAN
|
||||||
| NC score | 0.840176 (rank : 20) | θ value | 1.46607e-42 (rank : 23) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIK5_HUMAN
|
||||||
| NC score | 0.840139 (rank : 21) | θ value | 6.15952e-41 (rank : 26) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| NC score | 0.839710 (rank : 22) | θ value | 8.04462e-41 (rank : 27) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRID1_MOUSE
|
||||||
| NC score | 0.839067 (rank : 23) | θ value | 3.86705e-35 (rank : 31) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIA1_MOUSE
|
||||||
| NC score | 0.837557 (rank : 24) | θ value | 1.788e-48 (rank : 14) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRID1_HUMAN
|
||||||
| NC score | 0.837227 (rank : 25) | θ value | 2.12192e-33 (rank : 32) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIA1_HUMAN
|
||||||
| NC score | 0.836104 (rank : 26) | θ value | 4.40402e-47 (rank : 15) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA4_HUMAN
|
||||||
| NC score | 0.834166 (rank : 27) | θ value | 1.28137e-46 (rank : 16) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA4_MOUSE
|
||||||
| NC score | 0.832091 (rank : 28) | θ value | 2.18568e-46 (rank : 17) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA2_HUMAN
|
||||||
| NC score | 0.831727 (rank : 29) | θ value | 2.95404e-43 (rank : 21) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA2_MOUSE
|
||||||
| NC score | 0.831056 (rank : 30) | θ value | 8.59492e-43 (rank : 22) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA3_HUMAN
|
||||||
| NC score | 0.830906 (rank : 31) | θ value | 4.71619e-41 (rank : 25) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA3_MOUSE
|
||||||
| NC score | 0.830896 (rank : 32) | θ value | 3.99248e-40 (rank : 28) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIK1_MOUSE
|
||||||
| NC score | 0.823029 (rank : 33) | θ value | 2.34606e-32 (rank : 33) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
ERB1_HUMAN
|
||||||
| NC score | 0.136239 (rank : 34) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
TS1R1_MOUSE
|
||||||
| NC score | 0.062096 (rank : 35) | θ value | 0.00035302 (rank : 34) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.053306 (rank : 36) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.053173 (rank : 37) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
TS1R1_HUMAN
|
||||||
| NC score | 0.042524 (rank : 38) | θ value | 0.0193708 (rank : 35) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
DLEC1_MOUSE
|
||||||
| NC score | 0.036094 (rank : 39) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLA1 | Gene names | Dlec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in lung and esophageal cancer protein 1 homolog. | |||||
|
TS13_MOUSE
|
||||||
| NC score | 0.029590 (rank : 40) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
CHSP1_MOUSE
|
||||||
| NC score | 0.023758 (rank : 41) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CR86 | Gene names | Carhsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-regulated heat stable protein 1 (Calcium-regulated heat-stable protein of 24 kDa) (CRHSP-24). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.022614 (rank : 42) | θ value | 0.0961366 (rank : 37) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.022106 (rank : 43) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
EFS_MOUSE
|
||||||
| NC score | 0.021954 (rank : 44) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.021588 (rank : 45) | θ value | 0.0563607 (rank : 36) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.018380 (rank : 46) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
CK056_HUMAN
|
||||||
| NC score | 0.018054 (rank : 47) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N612, Q9C0A4, Q9H0N3, Q9H624 | Gene names | C11orf56, KIAA1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56. | |||||
|
CDT1_HUMAN
|
||||||
| NC score | 0.017165 (rank : 48) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.017108 (rank : 49) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.016198 (rank : 50) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RSPO4_MOUSE
|
||||||
| NC score | 0.015800 (rank : 51) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJ73 | Gene names | Rspo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (Cysteine-rich and single thrombospondin domain-containing protein 4) (Cristin-4) (mCristin-4). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.015722 (rank : 52) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
SHAN2_MOUSE
|
||||||
| NC score | 0.015526 (rank : 53) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.014502 (rank : 54) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
SHAN2_HUMAN
|
||||||
| NC score | 0.014439 (rank : 55) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.014317 (rank : 56) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CK056_MOUSE
|
||||||
| NC score | 0.013820 (rank : 57) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U2I3, Q3TWF5, Q6P543, Q80T93, Q811J6, Q8BI34, Q8VE55 | Gene names | Kiaa1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56 homolog. | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.013574 (rank : 58) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.013194 (rank : 59) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.012254 (rank : 60) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.011198 (rank : 61) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.010545 (rank : 62) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.010021 (rank : 63) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.009804 (rank : 64) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
ENW1_HUMAN
|
||||||
| NC score | 0.008696 (rank : 65) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQF0, O95244, O95245, Q8NHY7, Q9NRZ2, Q9NZG3 | Gene names | ERVWE1 | |||
|
Domain Architecture |

|
|||||
| Description | HERV-W_7q21.2 provirus ancestral Env polyprotein precursor (Envelope polyprotein gPr73) (HERV-7q Envelope protein) (HERV-W envelope protein) (Syncytin) (Syncytin-1) (Enverin) (Env-W) [Contains: Surface protein (SU) (gp50); Transmembrane protein (TM) (gp24)]. | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.008596 (rank : 66) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
TSKS_HUMAN
|
||||||
| NC score | 0.007842 (rank : 67) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJT2, Q8WXJ0 | Gene names | STK22S1, TSKS, TSKS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
TAF6L_MOUSE
|
||||||
| NC score | 0.007226 (rank : 68) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |

|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
VINEX_MOUSE
|
||||||
| NC score | 0.006544 (rank : 69) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.006413 (rank : 70) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TENS4_MOUSE
|
||||||
| NC score | 0.006397 (rank : 71) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.006362 (rank : 72) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.005922 (rank : 73) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NAB2_HUMAN
|
||||||
| NC score | 0.005625 (rank : 74) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15742, O76006, Q14797 | Gene names | NAB2, MADER | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2) (Melanoma- associated delayed early response protein) (Protein MADER). | |||||
|
LAMC3_HUMAN
|
||||||
| NC score | 0.004142 (rank : 75) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
BTBD7_HUMAN
|
||||||
| NC score | 0.003562 (rank : 76) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P203, Q69Z05, Q7Z308, Q86TS0, Q9HAA4, Q9NVM0 | Gene names | BTBD7, KIAA1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
CIAS1_MOUSE
|
||||||
| NC score | 0.002786 (rank : 77) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.002689 (rank : 78) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
MMP11_HUMAN
|
||||||
| NC score | 0.000759 (rank : 79) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
ADA25_MOUSE
|
||||||
| NC score | 0.000006 (rank : 80) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R159, Q9D4E4 | Gene names | Adam25 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 25 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 25) (Testase 2). | |||||
|
DLX5_HUMAN
|
||||||
| NC score | -0.000745 (rank : 81) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56178, Q9UPL1 | Gene names | DLX5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
DLX5_MOUSE
|
||||||
| NC score | -0.000805 (rank : 82) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
NRPN_MOUSE
|
||||||
| NC score | -0.001218 (rank : 83) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61955 | Gene names | Klk8, Nrpn, Prss19 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropsin precursor (EC 3.4.21.-) (NP) (Kallikrein-8). | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | -0.002026 (rank : 84) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
ZEP2_HUMAN
|
||||||
| NC score | -0.002067 (rank : 85) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
KCC2G_HUMAN
|
||||||
| NC score | -0.003310 (rank : 86) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13555, O15378, Q13556, Q5SQZ4 | Gene names | CAMK2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II gamma chain (EC 2.7.11.17) (CaM-kinase II gamma chain) (CaM kinase II gamma subunit) (CaMK-II subunit gamma). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | -0.003357 (rank : 87) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | -0.004104 (rank : 88) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||