Please be patient as the page loads
|
NMDE3_HUMAN
|
||||||
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NMDE1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.986307 (rank : 8) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMDE1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987121 (rank : 4) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.987058 (rank : 5) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.986861 (rank : 7) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.995986 (rank : 2) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.988702 (rank : 3) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE4_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.986900 (rank : 6) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMDZ1_MOUSE
|
||||||
| θ value | 2.73936e-73 (rank : 9) | NC score | 0.909670 (rank : 13) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_HUMAN
|
||||||
| θ value | 4.67263e-73 (rank : 10) | NC score | 0.910096 (rank : 12) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMD3A_HUMAN
|
||||||
| θ value | 2.17053e-70 (rank : 11) | NC score | 0.934014 (rank : 9) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMD3B_MOUSE
|
||||||
| θ value | 3.70236e-70 (rank : 12) | NC score | 0.927315 (rank : 10) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| θ value | 1.83746e-69 (rank : 13) | NC score | 0.926551 (rank : 11) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
GRIA1_MOUSE
|
||||||
| θ value | 1.97686e-47 (rank : 14) | NC score | 0.835807 (rank : 25) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_HUMAN
|
||||||
| θ value | 6.35935e-46 (rank : 15) | NC score | 0.834346 (rank : 26) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA4_HUMAN
|
||||||
| θ value | 8.30557e-46 (rank : 16) | NC score | 0.832390 (rank : 27) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA4_MOUSE
|
||||||
| θ value | 2.41655e-45 (rank : 17) | NC score | 0.830302 (rank : 28) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIK1_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 18) | NC score | 0.839973 (rank : 18) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIK4_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 19) | NC score | 0.840111 (rank : 17) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK4_MOUSE
|
||||||
| θ value | 2.95404e-43 (rank : 20) | NC score | 0.839872 (rank : 19) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIA2_HUMAN
|
||||||
| θ value | 1.91475e-42 (rank : 21) | NC score | 0.829960 (rank : 29) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIK2_HUMAN
|
||||||
| θ value | 2.50074e-42 (rank : 22) | NC score | 0.838591 (rank : 20) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIA2_MOUSE
|
||||||
| θ value | 5.57106e-42 (rank : 23) | NC score | 0.829285 (rank : 30) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIK3_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 24) | NC score | 0.840423 (rank : 16) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRIA3_HUMAN
|
||||||
| θ value | 3.05695e-40 (rank : 25) | NC score | 0.829129 (rank : 31) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIK5_HUMAN
|
||||||
| θ value | 3.05695e-40 (rank : 26) | NC score | 0.838540 (rank : 21) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| θ value | 3.99248e-40 (rank : 27) | NC score | 0.838072 (rank : 22) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIA3_MOUSE
|
||||||
| θ value | 1.16164e-39 (rank : 28) | NC score | 0.829127 (rank : 32) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRID2_MOUSE
|
||||||
| θ value | 3.16345e-37 (rank : 29) | NC score | 0.841839 (rank : 14) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID2_HUMAN
|
||||||
| θ value | 4.13158e-37 (rank : 30) | NC score | 0.840670 (rank : 15) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID1_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 31) | NC score | 0.837708 (rank : 23) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRID1_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 32) | NC score | 0.835862 (rank : 24) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK1_MOUSE
|
||||||
| θ value | 8.91499e-32 (rank : 33) | NC score | 0.821361 (rank : 33) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
TS1R1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 34) | NC score | 0.061636 (rank : 35) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
TS1R1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 35) | NC score | 0.042023 (rank : 38) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 0.21417 (rank : 36) | NC score | 0.021870 (rank : 44) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
ERB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.134967 (rank : 34) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
ASB10_HUMAN
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.007404 (rank : 78) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |
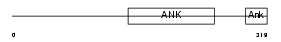
|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB10_MOUSE
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.007325 (rank : 79) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.029887 (rank : 39) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
TM55B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.018923 (rank : 47) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TWL2 | Gene names | Tmem55b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.016053 (rank : 53) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PROP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.008533 (rank : 74) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.024750 (rank : 41) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.013910 (rank : 63) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ATP4A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.017123 (rank : 48) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20648, O00738 | Gene names | ATP4A | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
CHSP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.025293 (rank : 40) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CR86 | Gene names | Carhsp1 | |||
|
Domain Architecture |
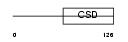
|
|||||
| Description | Calcium-regulated heat stable protein 1 (Calcium-regulated heat-stable protein of 24 kDa) (CRHSP-24). | |||||
|
DBND2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.020239 (rank : 45) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
Domain Architecture |
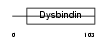
|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.014256 (rank : 62) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
ATP4A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.016830 (rank : 51) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64436, Q9CV46 | Gene names | Atp4a | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.008267 (rank : 75) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.011521 (rank : 66) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.023844 (rank : 42) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
AT1A4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.016371 (rank : 52) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.010804 (rank : 68) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.007805 (rank : 77) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
MAGD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.011384 (rank : 67) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.017015 (rank : 50) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.017094 (rank : 49) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.020194 (rank : 46) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.023156 (rank : 43) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.012805 (rank : 64) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
DOK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.008716 (rank : 72) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L591, Q8N864, Q9BQB3, Q9H666 | Gene names | DOK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.009758 (rank : 70) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.006827 (rank : 80) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
PRGC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.009863 (rank : 69) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.006679 (rank : 81) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.003386 (rank : 100) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
K1199_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.007855 (rank : 76) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BI06, Q6L9J4, Q9EPQ3, Q9EPQ4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1199 homolog precursor (Protein 12H19.01.T7). | |||||
|
AT1A2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.015478 (rank : 56) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.015464 (rank : 57) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
AT1A3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.015695 (rank : 55) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.015703 (rank : 54) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
ATS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.005215 (rank : 86) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHI8, Q9NSJ8, Q9P2K0, Q9UH83, Q9UP80 | Gene names | ADAMTS1, KIAA1346, METH1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1) (METH-1). | |||||
|
ATS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.005131 (rank : 87) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97857, O54768 | Gene names | Adamts1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.006426 (rank : 84) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.014437 (rank : 61) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CT174_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.011898 (rank : 65) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
MISS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.006461 (rank : 83) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
SLIT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | -0.002760 (rank : 109) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75093, Q8WWZ2, Q9UIL7 | Gene names | SLIT1, KIAA0813, MEGF4, SLIL1 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1) (Multiple epidermal growth factor-like domains 4). | |||||
|
WEE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | -0.003077 (rank : 110) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47810, Q9EPL7 | Gene names | Wee1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | -0.001578 (rank : 107) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
AT12A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.015241 (rank : 59) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z1W8, Q8VHY2 | Gene names | Atp12a, Atp1al1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT1A4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.015275 (rank : 58) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
ATS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.004553 (rank : 97) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
ATS5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.004772 (rank : 91) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R001 | Gene names | Adamts5 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-5 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 5) (ADAM-TS 5) (ADAM-TS5) (Aggrecanase-2) (ADMP-2) (Implantin). | |||||
|
AUXI_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.004661 (rank : 94) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.005118 (rank : 88) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CAN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.001624 (rank : 104) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.001075 (rank : 106) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
EFS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.008659 (rank : 73) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
FOXO4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.002547 (rank : 103) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |
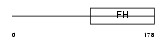
|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
KC1G1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | -0.002599 (rank : 108) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BTH8 | Gene names | Csnk1g1 | |||
|
Domain Architecture |
|
|||||
| Description | Casein kinase I isoform gamma-1 (EC 2.7.11.1) (CKI-gamma 1). | |||||
|
KR414_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.004723 (rank : 92) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KRA45_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.005074 (rank : 89) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
LBN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.003344 (rank : 101) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UK5, Q86YT3, Q86YT4, Q8NG49 | Gene names | EVC2, LBN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
M3K11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | -0.004669 (rank : 112) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.003687 (rank : 99) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
SON_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.009554 (rank : 71) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.005702 (rank : 85) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
XKR7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.004260 (rank : 98) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5GH72, Q9NUG5 | Gene names | XKR7, C20orf159, XRG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
ZBTB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | -0.003281 (rank : 111) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H5J0 | Gene names | ZBTB3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 3. | |||||
|
AT12A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.014941 (rank : 60) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.006666 (rank : 82) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CIAS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.002775 (rank : 102) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.004575 (rank : 96) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
CTGE4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.001408 (rank : 105) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IX94, O95046 | Gene names | CTAGE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 4 (cTAGE-4 protein). | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.004672 (rank : 93) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.004630 (rank : 95) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
K0460_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.005032 (rank : 90) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
GABR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.052940 (rank : 36) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.052808 (rank : 37) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_MOUSE
|
||||||
| NC score | 0.995986 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.988702 (rank : 3) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE1_MOUSE
|
||||||
| NC score | 0.987121 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| NC score | 0.987058 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE4_MOUSE
|
||||||
| NC score | 0.986900 (rank : 6) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMDE2_MOUSE
|
||||||
| NC score | 0.986861 (rank : 7) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE1_HUMAN
|
||||||
| NC score | 0.986307 (rank : 8) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMD3A_HUMAN
|
||||||
| NC score | 0.934014 (rank : 9) | θ value | 2.17053e-70 (rank : 11) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMD3B_MOUSE
|
||||||
| NC score | 0.927315 (rank : 10) | θ value | 3.70236e-70 (rank : 12) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| NC score | 0.926551 (rank : 11) | θ value | 1.83746e-69 (rank : 13) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
NMDZ1_HUMAN
|
||||||
| NC score | 0.910096 (rank : 12) | θ value | 4.67263e-73 (rank : 10) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| NC score | 0.909670 (rank : 13) | θ value | 2.73936e-73 (rank : 9) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
GRID2_MOUSE
|
||||||
| NC score | 0.841839 (rank : 14) | θ value | 3.16345e-37 (rank : 29) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID2_HUMAN
|
||||||
| NC score | 0.840670 (rank : 15) | θ value | 4.13158e-37 (rank : 30) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIK3_HUMAN
|
||||||
| NC score | 0.840423 (rank : 16) | θ value | 8.04462e-41 (rank : 24) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRIK4_HUMAN
|
||||||
| NC score | 0.840111 (rank : 17) | θ value | 1.73182e-43 (rank : 19) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK1_HUMAN
|
||||||
| NC score | 0.839973 (rank : 18) | θ value | 1.73182e-43 (rank : 18) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIK4_MOUSE
|
||||||
| NC score | 0.839872 (rank : 19) | θ value | 2.95404e-43 (rank : 20) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIK2_HUMAN
|
||||||
| NC score | 0.838591 (rank : 20) | θ value | 2.50074e-42 (rank : 22) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIK5_HUMAN
|
||||||
| NC score | 0.838540 (rank : 21) | θ value | 3.05695e-40 (rank : 26) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| NC score | 0.838072 (rank : 22) | θ value | 3.99248e-40 (rank : 27) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRID1_MOUSE
|
||||||
| NC score | 0.837708 (rank : 23) | θ value | 1.12514e-34 (rank : 31) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRID1_HUMAN
|
||||||
| NC score | 0.835862 (rank : 24) | θ value | 6.17384e-33 (rank : 32) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIA1_MOUSE
|
||||||
| NC score | 0.835807 (rank : 25) | θ value | 1.97686e-47 (rank : 14) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_HUMAN
|
||||||
| NC score | 0.834346 (rank : 26) | θ value | 6.35935e-46 (rank : 15) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA4_HUMAN
|
||||||
| NC score | 0.832390 (rank : 27) | θ value | 8.30557e-46 (rank : 16) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA4_MOUSE
|
||||||
| NC score | 0.830302 (rank : 28) | θ value | 2.41655e-45 (rank : 17) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA2_HUMAN
|
||||||
| NC score | 0.829960 (rank : 29) | θ value | 1.91475e-42 (rank : 21) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA2_MOUSE
|
||||||
| NC score | 0.829285 (rank : 30) | θ value | 5.57106e-42 (rank : 23) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA3_HUMAN
|
||||||
| NC score | 0.829129 (rank : 31) | θ value | 3.05695e-40 (rank : 25) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA3_MOUSE
|
||||||
| NC score | 0.829127 (rank : 32) | θ value | 1.16164e-39 (rank : 28) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIK1_MOUSE
|
||||||
| NC score | 0.821361 (rank : 33) | θ value | 8.91499e-32 (rank : 33) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
ERB1_HUMAN
|
||||||
| NC score | 0.134967 (rank : 34) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
TS1R1_MOUSE
|
||||||
| NC score | 0.061636 (rank : 35) | θ value | 0.000461057 (rank : 34) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.052940 (rank : 36) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.052808 (rank : 37) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
TS1R1_HUMAN
|
||||||
| NC score | 0.042023 (rank : 38) | θ value | 0.0330416 (rank : 35) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.029887 (rank : 39) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CHSP1_MOUSE
|
||||||
| NC score | 0.025293 (rank : 40) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CR86 | Gene names | Carhsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-regulated heat stable protein 1 (Calcium-regulated heat-stable protein of 24 kDa) (CRHSP-24). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.024750 (rank : 41) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.023844 (rank : 42) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.023156 (rank : 43) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.021870 (rank : 44) | θ value | 0.21417 (rank : 36) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
DBND2_HUMAN
|
||||||
| NC score | 0.020239 (rank : 45) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
Domain Architecture |

|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.020194 (rank : 46) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TM55B_MOUSE
|
||||||
| NC score | 0.018923 (rank : 47) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TWL2 | Gene names | Tmem55b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
|
ATP4A_HUMAN
|
||||||
| NC score | 0.017123 (rank : 48) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20648, O00738 | Gene names | ATP4A | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.017094 (rank : 49) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.017015 (rank : 50) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
ATP4A_MOUSE
|
||||||
| NC score | 0.016830 (rank : 51) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64436, Q9CV46 | Gene names | Atp4a | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT1A4_MOUSE
|
||||||
| NC score | 0.016371 (rank : 52) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.016053 (rank : 53) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
AT1A3_MOUSE
|
||||||
| NC score | 0.015703 (rank : 54) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_HUMAN
|
||||||
| NC score | 0.015695 (rank : 55) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A2_HUMAN
|
||||||
| NC score | 0.015478 (rank : 56) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A2_MOUSE
|
||||||
| NC score | 0.015464 (rank : 57) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
AT1A4_HUMAN
|
||||||
| NC score | 0.015275 (rank : 58) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT12A_MOUSE
|
||||||
| NC score | 0.015241 (rank : 59) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z1W8, Q8VHY2 | Gene names | Atp12a, Atp1al1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT12A_HUMAN
|
||||||
| NC score | 0.014941 (rank : 60) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.014437 (rank : 61) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.014256 (rank : 62) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.013910 (rank : 63) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.012805 (rank : 64) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.011898 (rank : 65) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.011521 (rank : 66) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.011384 (rank : 67) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.010804 (rank : 68) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
PRGC2_HUMAN
|
||||||
| NC score | 0.009863 (rank : 69) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.009758 (rank : 70) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.009554 (rank : 71) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
DOK3_HUMAN
|
||||||
| NC score | 0.008716 (rank : 72) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L591, Q8N864, Q9BQB3, Q9H666 | Gene names | DOK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.008659 (rank : 73) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
PROP_MOUSE
|
||||||
| NC score | 0.008533 (rank : 74) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.008267 (rank : 75) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
K1199_MOUSE
|
||||||
| NC score | 0.007855 (rank : 76) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BI06, Q6L9J4, Q9EPQ3, Q9EPQ4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1199 homolog precursor (Protein 12H19.01.T7). | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.007805 (rank : 77) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
ASB10_HUMAN
|
||||||
| NC score | 0.007404 (rank : 78) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB10_MOUSE
|
||||||
| NC score | 0.007325 (rank : 79) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.006827 (rank : 80) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.006679 (rank : 81) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.006666 (rank : 82) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.006461 (rank : 83) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.006426 (rank : 84) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.005702 (rank : 85) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
ATS1_HUMAN
|
||||||
| NC score | 0.005215 (rank : 86) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHI8, Q9NSJ8, Q9P2K0, Q9UH83, Q9UP80 | Gene names | ADAMTS1, KIAA1346, METH1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1) (METH-1). | |||||
|
ATS1_MOUSE
|
||||||
| NC score | 0.005131 (rank : 87) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97857, O54768 | Gene names | Adamts1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.005118 (rank : 88) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
KRA45_HUMAN
|
||||||
| NC score | 0.005074 (rank : 89) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.005032 (rank : 90) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
ATS5_MOUSE
|
||||||
| NC score | 0.004772 (rank : 91) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R001 | Gene names | Adamts5 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-5 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 5) (ADAM-TS 5) (ADAM-TS5) (Aggrecanase-2) (ADMP-2) (Implantin). | |||||
|
KR414_HUMAN
|
||||||
| NC score | 0.004723 (rank : 92) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.004672 (rank : 93) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
AUXI_MOUSE
|
||||||
| NC score | 0.004661 (rank : 94) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.004630 (rank : 95) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.004575 (rank : 96) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
ATS2_HUMAN
|
||||||
| NC score | 0.004553 (rank : 97) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
XKR7_HUMAN
|
||||||
| NC score | 0.004260 (rank : 98) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5GH72, Q9NUG5 | Gene names | XKR7, C20orf159, XRG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.003687 (rank : 99) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.003386 (rank : 100) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
LBN_HUMAN
|
||||||
| NC score | 0.003344 (rank : 101) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UK5, Q86YT3, Q86YT4, Q8NG49 | Gene names | EVC2, LBN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
CIAS1_MOUSE
|
||||||
| NC score | 0.002775 (rank : 102) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
FOXO4_MOUSE
|
||||||
| NC score | 0.002547 (rank : 103) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
CAN3_MOUSE
|
||||||
| NC score | 0.001624 (rank : 104) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CTGE4_HUMAN
|
||||||
| NC score | 0.001408 (rank : 105) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IX94, O95046 | Gene names | CTAGE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 4 (cTAGE-4 protein). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.001075 (rank : 106) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
ZN516_MOUSE
|
||||||
| NC score | -0.001578 (rank : 107) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
KC1G1_MOUSE
|
||||||
| NC score | -0.002599 (rank : 108) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BTH8 | Gene names | Csnk1g1 | |||
|
Domain Architecture |

|
|||||
| Description | Casein kinase I isoform gamma-1 (EC 2.7.11.1) (CKI-gamma 1). | |||||
|
SLIT1_HUMAN
|
||||||
| NC score | -0.002760 (rank : 109) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75093, Q8WWZ2, Q9UIL7 | Gene names | SLIT1, KIAA0813, MEGF4, SLIL1 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1) (Multiple epidermal growth factor-like domains 4). | |||||
|
WEE1_MOUSE
|
||||||
| NC score | -0.003077 (rank : 110) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47810, Q9EPL7 | Gene names | Wee1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase). | |||||
|
ZBTB3_HUMAN
|
||||||
| NC score | -0.003281 (rank : 111) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H5J0 | Gene names | ZBTB3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 3. | |||||
|
M3K11_HUMAN
|
||||||
| NC score | -0.004669 (rank : 112) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||