Please be patient as the page loads
|
DBND2_HUMAN
|
||||||
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
Domain Architecture |
|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DBND2_HUMAN
|
||||||
| θ value | 2.9938e-112 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
Domain Architecture |

|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
|
DBND2_MOUSE
|
||||||
| θ value | 2.67181e-44 (rank : 2) | NC score | 0.961593 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CRD4 | Gene names | Dbndd2, D2Bwg0891e | |||
|
Domain Architecture |
|
|||||
| Description | Dysbindin domain-containing protein 2. | |||||
|
DTBP1_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 3) | NC score | 0.629977 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EV8, Q96NV2, Q9H0U2, Q9H3J5 | Gene names | DTNBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein). | |||||
|
DTBP1_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 4) | NC score | 0.618548 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WZ8, Q80ZN4 | Gene names | Dtnbp1 | |||
|
Domain Architecture |
|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein homolog). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.041471 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SMN_HUMAN
|
||||||
| θ value | 0.125558 (rank : 6) | NC score | 0.100580 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16637, Q13119, Q96J51 | Gene names | SMN1, SMN, SMNT | |||
|
Domain Architecture |
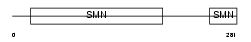
|
|||||
| Description | Survival motor neuron protein (Component of gems 1) (Gemin-1). | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.074203 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
IDD_MOUSE
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.077041 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |
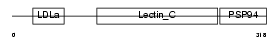
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.020239 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.061401 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.062512 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
ZN598_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.043799 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
CAD22_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.016342 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJ99, O43205 | Gene names | CDH22, C20orf25 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
CAD22_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.016257 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTP5 | Gene names | Cdh22 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.065553 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.024509 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
M3K11_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.004966 (rank : 43) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||
|
DAAM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.063934 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.060089 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.039468 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
DAAM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.062466 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
DIAP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.068647 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.032834 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.035598 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
NETR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.006138 (rank : 42) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.012927 (rank : 40) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.020975 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CCD1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.025563 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.060317 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.006318 (rank : 41) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.030216 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
WASP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.018565 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.013313 (rank : 39) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
K0256_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.017278 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6A098 | Gene names | Kiaa0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
OAS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.015037 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
|
SMN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.062718 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97801, O09092 | Gene names | Smn1, Smn | |||
|
Domain Architecture |
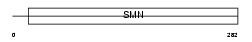
|
|||||
| Description | Survival motor neuron protein. | |||||
|
SON_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.025128 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TNR8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.016417 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |
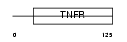
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
DAAM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.050951 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
DAAM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.051773 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.059390 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
DIAP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.056150 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSV4, Q5T2S7, Q5XKF6, Q6MZF0, Q6NUP0, Q86VS4, Q8NAV4 | Gene names | DIAPH3, DIAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3). | |||||
|
DIAP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.056068 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z207 | Gene names | Diaph3, Diap3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3) (mDIA2) (p134mDIA2). | |||||
|
DBND2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.9938e-112 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
Domain Architecture |

|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
|
DBND2_MOUSE
|
||||||
| NC score | 0.961593 (rank : 2) | θ value | 2.67181e-44 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CRD4 | Gene names | Dbndd2, D2Bwg0891e | |||
|
Domain Architecture |

|
|||||
| Description | Dysbindin domain-containing protein 2. | |||||
|
DTBP1_HUMAN
|
||||||
| NC score | 0.629977 (rank : 3) | θ value | 3.28887e-18 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EV8, Q96NV2, Q9H0U2, Q9H3J5 | Gene names | DTNBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein). | |||||
|
DTBP1_MOUSE
|
||||||
| NC score | 0.618548 (rank : 4) | θ value | 8.10077e-17 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WZ8, Q80ZN4 | Gene names | Dtnbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein homolog). | |||||
|
SMN_HUMAN
|
||||||
| NC score | 0.100580 (rank : 5) | θ value | 0.125558 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16637, Q13119, Q96J51 | Gene names | SMN1, SMN, SMNT | |||
|
Domain Architecture |

|
|||||
| Description | Survival motor neuron protein (Component of gems 1) (Gemin-1). | |||||
|
IDD_MOUSE
|
||||||
| NC score | 0.077041 (rank : 6) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.074203 (rank : 7) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
DIAP2_MOUSE
|
||||||
| NC score | 0.068647 (rank : 8) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.065553 (rank : 9) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
DAAM1_MOUSE
|
||||||
| NC score | 0.063934 (rank : 10) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
SMN_MOUSE
|
||||||
| NC score | 0.062718 (rank : 11) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97801, O09092 | Gene names | Smn1, Smn | |||
|
Domain Architecture |

|
|||||
| Description | Survival motor neuron protein. | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.062512 (rank : 12) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
DAAM1_HUMAN
|
||||||
| NC score | 0.062466 (rank : 13) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.061401 (rank : 14) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.060317 (rank : 15) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.060089 (rank : 16) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.059390 (rank : 17) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
DIAP3_HUMAN
|
||||||
| NC score | 0.056150 (rank : 18) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSV4, Q5T2S7, Q5XKF6, Q6MZF0, Q6NUP0, Q86VS4, Q8NAV4 | Gene names | DIAPH3, DIAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3). | |||||
|
DIAP3_MOUSE
|
||||||
| NC score | 0.056068 (rank : 19) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z207 | Gene names | Diaph3, Diap3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3) (mDIA2) (p134mDIA2). | |||||
|
DAAM2_MOUSE
|
||||||
| NC score | 0.051773 (rank : 20) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
DAAM2_HUMAN
|
||||||
| NC score | 0.050951 (rank : 21) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.043799 (rank : 22) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.041471 (rank : 23) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.039468 (rank : 24) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.035598 (rank : 25) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.032834 (rank : 26) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.030216 (rank : 27) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
CCD1A_HUMAN
|
||||||
| NC score | 0.025563 (rank : 28) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.025128 (rank : 29) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.024509 (rank : 30) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.020975 (rank : 31) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.020239 (rank : 32) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.018565 (rank : 33) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
K0256_MOUSE
|
||||||
| NC score | 0.017278 (rank : 34) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6A098 | Gene names | Kiaa0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
TNR8_MOUSE
|
||||||
| NC score | 0.016417 (rank : 35) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
CAD22_HUMAN
|
||||||
| NC score | 0.016342 (rank : 36) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJ99, O43205 | Gene names | CDH22, C20orf25 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
CAD22_MOUSE
|
||||||
| NC score | 0.016257 (rank : 37) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTP5 | Gene names | Cdh22 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
OAS3_HUMAN
|
||||||
| NC score | 0.015037 (rank : 38) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.013313 (rank : 39) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.012927 (rank : 40) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.006318 (rank : 41) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
NETR_HUMAN
|
||||||
| NC score | 0.006138 (rank : 42) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
M3K11_HUMAN
|
||||||
| NC score | 0.004966 (rank : 43) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||