Please be patient as the page loads
|
MAGD1_MOUSE
|
||||||
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MAGD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.979967 (rank : 2) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MAGD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 2.13557e-126 (rank : 3) | NC score | 0.942741 (rank : 7) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
TROP_HUMAN
|
||||||
| θ value | 2.15048e-102 (rank : 4) | NC score | 0.933460 (rank : 9) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
MAGD2_HUMAN
|
||||||
| θ value | 6.48998e-91 (rank : 5) | NC score | 0.968491 (rank : 3) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MAGL2_HUMAN
|
||||||
| θ value | 2.041e-52 (rank : 6) | NC score | 0.941577 (rank : 8) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
MAGG1_HUMAN
|
||||||
| θ value | 9.48078e-50 (rank : 7) | NC score | 0.946702 (rank : 5) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96MG7, Q8IW16, Q8TEI6, Q9H214 | Gene names | NDNL2, HCA4, MAGEG1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2) (Hepatocellular carcinoma-associated protein 4). | |||||
|
MAGG1_MOUSE
|
||||||
| θ value | 2.1121e-49 (rank : 8) | NC score | 0.947377 (rank : 4) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 9) | NC score | 0.665990 (rank : 36) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAGL2_MOUSE
|
||||||
| θ value | 1.01529e-43 (rank : 10) | NC score | 0.932502 (rank : 11) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QZ04 | Gene names | Magel2 | |||
|
Domain Architecture |
|
|||||
| Description | MAGE-like protein 2 (Protein nS7). | |||||
|
MAGB3_HUMAN
|
||||||
| θ value | 5.03882e-43 (rank : 11) | NC score | 0.927555 (rank : 13) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
MAGAB_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 12) | NC score | 0.908228 (rank : 25) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |
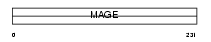
|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAGBA_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 13) | NC score | 0.929144 (rank : 12) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96LZ2, Q494Y6, Q494Y7, Q9BZ78 | Gene names | MAGEB10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B10 (MAGE-B10 antigen). | |||||
|
MAGB4_HUMAN
|
||||||
| θ value | 3.05695e-40 (rank : 14) | NC score | 0.924165 (rank : 17) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |
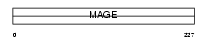
|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAGBI_HUMAN
|
||||||
| θ value | 7.52953e-39 (rank : 15) | NC score | 0.921175 (rank : 18) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
MAGF1_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 16) | NC score | 0.945651 (rank : 6) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAY2, Q9H215 | Gene names | MAGEF1 | |||
|
Domain Architecture |
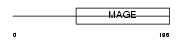
|
|||||
| Description | Melanoma-associated antigen F1 (MAGE-F1 antigen). | |||||
|
MAGB1_HUMAN
|
||||||
| θ value | 1.08726e-37 (rank : 17) | NC score | 0.920436 (rank : 19) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |
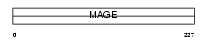
|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 2.67802e-36 (rank : 18) | NC score | 0.919243 (rank : 21) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MAGA1_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 19) | NC score | 0.896748 (rank : 27) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43355, O00346 | Gene names | MAGEA1, MAGE1, MAGE1A | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 1 (MAGE-1 antigen) (Antigen MZ2-E). | |||||
|
MAGB2_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 20) | NC score | 0.924928 (rank : 15) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
MAGAA_HUMAN
|
||||||
| θ value | 2.26708e-35 (rank : 21) | NC score | 0.908315 (rank : 24) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
NECD_HUMAN
|
||||||
| θ value | 2.26708e-35 (rank : 22) | NC score | 0.932711 (rank : 10) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |
|
|||||
| Description | Necdin. | |||||
|
MAGE2_HUMAN
|
||||||
| θ value | 8.61488e-35 (rank : 23) | NC score | 0.924831 (rank : 16) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TD90 | Gene names | MAGEE2, HCA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E2 (MAGE-E2 antigen) (Hepatocellular carcinoma-associated protein 3). | |||||
|
MAGA4_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 24) | NC score | 0.892005 (rank : 28) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43358, Q14798 | Gene names | MAGEA4, MAGE4 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 4 (MAGE-4 antigen) (MAGE-X2) (MAGE-41). | |||||
|
NECD_MOUSE
|
||||||
| θ value | 5.584e-34 (rank : 25) | NC score | 0.926797 (rank : 14) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MAGA6_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 26) | NC score | 0.880959 (rank : 29) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43360 | Gene names | MAGEA6, MAGE6 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 6 (MAGE-6 antigen) (MAGE3B). | |||||
|
MAGAC_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 27) | NC score | 0.880349 (rank : 30) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 28) | NC score | 0.863207 (rank : 33) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAGA9_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 29) | NC score | 0.899864 (rank : 26) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43362, Q92910 | Gene names | MAGEA9, MAGE9 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 9 (MAGE-9 antigen). | |||||
|
MAGC2_HUMAN
|
||||||
| θ value | 3.17079e-29 (rank : 30) | NC score | 0.912624 (rank : 23) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UBF1, Q5JZ00, Q96D45, Q9P1M6, Q9P1M7 | Gene names | MAGEC2, HCA587, MAGEE1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen C2 (MAGE-C2 antigen) (MAGE-E1 antigen) (Hepatocellular carcinoma-associated antigen 587) (Cancer-testis antigen 10) (CT10). | |||||
|
MAGA3_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 31) | NC score | 0.878016 (rank : 31) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43357 | Gene names | MAGEA3, MAGE3 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 3 (MAGE-3 antigen) (Antigen MZ2-D). | |||||
|
MAGH1_MOUSE
|
||||||
| θ value | 1.92365e-26 (rank : 32) | NC score | 0.918255 (rank : 22) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NWG9, Q99PB0 | Gene names | Mageh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen). | |||||
|
MAGA2_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 33) | NC score | 0.876688 (rank : 32) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43356 | Gene names | MAGEA2, MAGE2, MAGEA2A | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 2 (MAGE-2 antigen). | |||||
|
MAGH1_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 34) | NC score | 0.919285 (rank : 20) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H213, Q5JRJ3, Q9Y5M2 | Gene names | MAGEH1, APR1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen) (Restin) (Apoptosis- related protein 1) (APR-1). | |||||
|
MAGC3_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 35) | NC score | 0.846218 (rank : 35) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
MAGA8_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 36) | NC score | 0.847042 (rank : 34) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 37) | NC score | 0.047698 (rank : 74) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 38) | NC score | 0.041662 (rank : 84) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 39) | NC score | 0.074182 (rank : 47) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
RBM14_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 40) | NC score | 0.044434 (rank : 78) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
RBM14_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 41) | NC score | 0.044394 (rank : 79) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 42) | NC score | 0.075935 (rank : 46) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 43) | NC score | 0.084898 (rank : 40) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 44) | NC score | 0.049638 (rank : 72) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 45) | NC score | 0.066001 (rank : 53) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 46) | NC score | 0.072094 (rank : 48) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 47) | NC score | 0.043158 (rank : 82) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.025649 (rank : 106) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 49) | NC score | 0.044313 (rank : 80) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 50) | NC score | 0.080321 (rank : 42) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 51) | NC score | 0.023449 (rank : 111) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 52) | NC score | 0.052754 (rank : 67) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 53) | NC score | 0.052337 (rank : 69) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HXD12_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 54) | NC score | 0.007138 (rank : 141) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23812 | Gene names | Hoxd12, Hox-4.7, Hoxd-12 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D12 (Hox-4.7) (Hox-5.6). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 55) | NC score | 0.037180 (rank : 90) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
CASL_HUMAN
|
||||||
| θ value | 0.279714 (rank : 56) | NC score | 0.022462 (rank : 112) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |
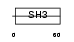
|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 57) | NC score | 0.056041 (rank : 64) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.365318 (rank : 58) | NC score | 0.070698 (rank : 50) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.365318 (rank : 59) | NC score | 0.044708 (rank : 77) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.365318 (rank : 60) | NC score | 0.039844 (rank : 86) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SCYL2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 61) | NC score | 0.007611 (rank : 139) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 632 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3W7, Q96EF4, Q96ST4, Q9H7V5, Q9NVH3, Q9P2I7 | Gene names | SCYL2, CVAK104, KIAA1360 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
P80C_HUMAN
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.032216 (rank : 96) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.053524 (rank : 66) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
CENPC_MOUSE
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.029754 (rank : 99) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49452 | Gene names | Cenpc1, Cenpc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C). | |||||
|
DSRAD_MOUSE
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.015902 (rank : 124) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.026109 (rank : 105) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.62314 (rank : 67) | NC score | 0.035372 (rank : 91) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.037505 (rank : 89) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 69) | NC score | 0.006512 (rank : 142) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.62314 (rank : 70) | NC score | 0.070952 (rank : 49) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
ZCC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 71) | NC score | 0.019583 (rank : 113) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |
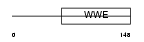
|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.048987 (rank : 73) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
CNTN6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.008935 (rank : 134) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JMB8, Q8C6X1 | Gene names | Cntn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (mNB-3). | |||||
|
NCOA5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 74) | NC score | 0.027630 (rank : 103) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91W39 | Gene names | Ncoa5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 0.813845 (rank : 75) | NC score | 0.079363 (rank : 44) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
CNTN3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.008044 (rank : 138) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P232, Q9H039 | Gene names | CNTN3, KIAA1496, PANG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 77) | NC score | 0.016343 (rank : 122) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 78) | NC score | 0.032830 (rank : 95) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.017864 (rank : 119) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 80) | NC score | 0.011563 (rank : 128) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.040752 (rank : 85) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
CNTN6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.008655 (rank : 135) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ52 | Gene names | CNTN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (hNB-3). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.042175 (rank : 83) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TAB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.018208 (rank : 118) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYJ8, O94838, Q6I9W8, Q76N06, Q9UFP7 | Gene names | MAP3K7IP2, KIAA0733, TAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.044180 (rank : 81) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.017782 (rank : 120) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.038197 (rank : 88) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.025241 (rank : 107) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.034769 (rank : 93) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.025059 (rank : 108) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.047047 (rank : 75) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.047016 (rank : 76) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
YTHD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.018679 (rank : 117) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.012043 (rank : 127) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZN579_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | -0.003649 (rank : 152) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NAF0 | Gene names | ZNF579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 579. | |||||
|
CF010_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.034996 (rank : 92) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.003912 (rank : 146) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
MARK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | -0.001815 (rank : 150) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.011017 (rank : 130) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.055701 (rank : 65) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NOL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.019399 (rank : 114) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.027979 (rank : 102) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
BASP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.108049 (rank : 38) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
EMID2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.010127 (rank : 133) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96A83 | Gene names | EMID2, COL26A1, EMU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
FUS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.024185 (rank : 110) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
MRP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.030532 (rank : 97) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
PAWR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.019062 (rank : 116) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.029212 (rank : 100) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
YTHD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.013235 (rank : 126) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59326, Q3T9E2 | Gene names | Ythdf1 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1 homolog) (DACA-1 homolog). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.001203 (rank : 149) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CNTN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.007284 (rank : 140) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07409 | Gene names | Cntn3, Pang, Pcs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.033852 (rank : 94) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.024403 (rank : 109) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MUCEN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.027581 (rank : 104) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
PALM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.016184 (rank : 123) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
PHAR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.028283 (rank : 101) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.008370 (rank : 137) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.060469 (rank : 57) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.039721 (rank : 87) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.014665 (rank : 125) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
TSYL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.017091 (rank : 121) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88852, Q3TKW0 | Gene names | Tspyl1, Tspyl | |||
|
Domain Architecture |
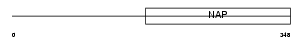
|
|||||
| Description | Testis-specific Y-encoded-like protein 1 (TSPY-like 1). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.010419 (rank : 132) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | -0.003490 (rank : 151) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
COCA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.004534 (rank : 144) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
K1949_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.019320 (rank : 115) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
MIME_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.001295 (rank : 148) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20774, Q9UF90, Q9UNK5 | Gene names | OGN, OIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mimecan precursor (Osteoglycin) (Osteoinductive factor) (OIF). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.011519 (rank : 129) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.050790 (rank : 71) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.008650 (rank : 136) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.030510 (rank : 98) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CNTN4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.006405 (rank : 143) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q69Z26, Q8BRT6, Q8CBD3 | Gene names | Cntn4, Kiaa3024 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
ERBB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | -0.004267 (rank : 153) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.010927 (rank : 131) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.065848 (rank : 54) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.001718 (rank : 147) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
SHE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.004038 (rank : 145) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BSD5, Q3TZT0 | Gene names | She | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein E. | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.067754 (rank : 52) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.052341 (rank : 68) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.076994 (rank : 45) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.056210 (rank : 63) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
LORI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.103500 (rank : 39) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LORI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.068796 (rank : 51) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
MAGA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.466214 (rank : 37) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P43359 | Gene names | MAGEA5, MAGE5 | |||
|
Domain Architecture |
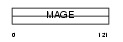
|
|||||
| Description | Melanoma-associated antigen 5 (MAGE-5 antigen). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.084011 (rank : 41) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.056540 (rank : 61) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.060298 (rank : 58) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.052166 (rank : 70) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
PHXR5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.079999 (rank : 43) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
PO121_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.060136 (rank : 59) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.059020 (rank : 60) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.065373 (rank : 56) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SELPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.065424 (rank : 55) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.056297 (rank : 62) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
MAGD1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.979967 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MAGD2_HUMAN
|
||||||
| NC score | 0.968491 (rank : 3) | θ value | 6.48998e-91 (rank : 5) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MAGG1_MOUSE
|
||||||
| NC score | 0.947377 (rank : 4) | θ value | 2.1121e-49 (rank : 8) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
MAGG1_HUMAN
|
||||||
| NC score | 0.946702 (rank : 5) | θ value | 9.48078e-50 (rank : 7) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96MG7, Q8IW16, Q8TEI6, Q9H214 | Gene names | NDNL2, HCA4, MAGEG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2) (Hepatocellular carcinoma-associated protein 4). | |||||
|
MAGF1_HUMAN
|
||||||
| NC score | 0.945651 (rank : 6) | θ value | 2.86122e-38 (rank : 16) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAY2, Q9H215 | Gene names | MAGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen F1 (MAGE-F1 antigen). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.942741 (rank : 7) | θ value | 2.13557e-126 (rank : 3) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
MAGL2_HUMAN
|
||||||
| NC score | 0.941577 (rank : 8) | θ value | 2.041e-52 (rank : 6) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
TROP_HUMAN
|
||||||
| NC score | 0.933460 (rank : 9) | θ value | 2.15048e-102 (rank : 4) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
NECD_HUMAN
|
||||||
| NC score | 0.932711 (rank : 10) | θ value | 2.26708e-35 (rank : 22) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MAGL2_MOUSE
|
||||||
| NC score | 0.932502 (rank : 11) | θ value | 1.01529e-43 (rank : 10) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QZ04 | Gene names | Magel2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Protein nS7). | |||||
|
MAGBA_HUMAN
|
||||||
| NC score | 0.929144 (rank : 12) | θ value | 2.11701e-41 (rank : 13) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96LZ2, Q494Y6, Q494Y7, Q9BZ78 | Gene names | MAGEB10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B10 (MAGE-B10 antigen). | |||||
|
MAGB3_HUMAN
|
||||||
| NC score | 0.927555 (rank : 13) | θ value | 5.03882e-43 (rank : 11) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
NECD_MOUSE
|
||||||
| NC score | 0.926797 (rank : 14) | θ value | 5.584e-34 (rank : 25) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MAGB2_HUMAN
|
||||||
| NC score | 0.924928 (rank : 15) | θ value | 5.96599e-36 (rank : 20) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
MAGE2_HUMAN
|
||||||
| NC score | 0.924831 (rank : 16) | θ value | 8.61488e-35 (rank : 23) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TD90 | Gene names | MAGEE2, HCA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E2 (MAGE-E2 antigen) (Hepatocellular carcinoma-associated protein 3). | |||||
|
MAGB4_HUMAN
|
||||||
| NC score | 0.924165 (rank : 17) | θ value | 3.05695e-40 (rank : 14) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAGBI_HUMAN
|
||||||
| NC score | 0.921175 (rank : 18) | θ value | 7.52953e-39 (rank : 15) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
MAGB1_HUMAN
|
||||||
| NC score | 0.920436 (rank : 19) | θ value | 1.08726e-37 (rank : 17) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
MAGH1_HUMAN
|
||||||
| NC score | 0.919285 (rank : 20) | θ value | 5.59698e-26 (rank : 34) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H213, Q5JRJ3, Q9Y5M2 | Gene names | MAGEH1, APR1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen) (Restin) (Apoptosis- related protein 1) (APR-1). | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.919243 (rank : 21) | θ value | 2.67802e-36 (rank : 18) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MAGH1_MOUSE
|
||||||
| NC score | 0.918255 (rank : 22) | θ value | 1.92365e-26 (rank : 32) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NWG9, Q99PB0 | Gene names | Mageh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen). | |||||
|
MAGC2_HUMAN
|
||||||
| NC score | 0.912624 (rank : 23) | θ value | 3.17079e-29 (rank : 30) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UBF1, Q5JZ00, Q96D45, Q9P1M6, Q9P1M7 | Gene names | MAGEC2, HCA587, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C2 (MAGE-C2 antigen) (MAGE-E1 antigen) (Hepatocellular carcinoma-associated antigen 587) (Cancer-testis antigen 10) (CT10). | |||||
|
MAGAA_HUMAN
|
||||||
| NC score | 0.908315 (rank : 24) | θ value | 2.26708e-35 (rank : 21) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
MAGAB_HUMAN
|
||||||
| NC score | 0.908228 (rank : 25) | θ value | 2.11701e-41 (rank : 12) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAGA9_HUMAN
|
||||||
| NC score | 0.899864 (rank : 26) | θ value | 2.86786e-30 (rank : 29) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43362, Q92910 | Gene names | MAGEA9, MAGE9 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 9 (MAGE-9 antigen). | |||||
|
MAGA1_HUMAN
|
||||||
| NC score | 0.896748 (rank : 27) | θ value | 5.96599e-36 (rank : 19) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43355, O00346 | Gene names | MAGEA1, MAGE1, MAGE1A | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 1 (MAGE-1 antigen) (Antigen MZ2-E). | |||||
|
MAGA4_HUMAN
|
||||||
| NC score | 0.892005 (rank : 28) | θ value | 1.12514e-34 (rank : 24) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43358, Q14798 | Gene names | MAGEA4, MAGE4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 4 (MAGE-4 antigen) (MAGE-X2) (MAGE-41). | |||||
|
MAGA6_HUMAN
|
||||||
| NC score | 0.880959 (rank : 29) | θ value | 1.52067e-31 (rank : 26) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43360 | Gene names | MAGEA6, MAGE6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 6 (MAGE-6 antigen) (MAGE3B). | |||||
|
MAGAC_HUMAN
|
||||||
| NC score | 0.880349 (rank : 30) | θ value | 1.52067e-31 (rank : 27) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
MAGA3_HUMAN
|
||||||
| NC score | 0.878016 (rank : 31) | θ value | 1.33217e-27 (rank : 31) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43357 | Gene names | MAGEA3, MAGE3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 3 (MAGE-3 antigen) (Antigen MZ2-D). | |||||
|
MAGA2_HUMAN
|
||||||
| NC score | 0.876688 (rank : 32) | θ value | 4.28545e-26 (rank : 33) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43356 | Gene names | MAGEA2, MAGE2, MAGEA2A | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 2 (MAGE-2 antigen). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.863207 (rank : 33) | θ value | 1.52067e-31 (rank : 28) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAGA8_HUMAN
|
||||||
| NC score | 0.847042 (rank : 34) | θ value | 2.35696e-16 (rank : 36) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
MAGC3_HUMAN
|
||||||
| NC score | 0.846218 (rank : 35) | θ value | 5.60996e-18 (rank : 35) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.665990 (rank : 36) | θ value | 2.3352e-48 (rank : 9) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAGA5_HUMAN
|
||||||
| NC score | 0.466214 (rank : 37) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P43359 | Gene names | MAGEA5, MAGE5 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 5 (MAGE-5 antigen). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.108049 (rank : 38) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.103500 (rank : 39) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.084898 (rank : 40) | θ value | 0.0148317 (rank : 43) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.084011 (rank : 41) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.080321 (rank : 42) | θ value | 0.0563607 (rank : 50) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PHXR5_MOUSE
|
||||||
| NC score | 0.079999 (rank : 43) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.079363 (rank : 44) | θ value | 0.813845 (rank : 75) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.076994 (rank : 45) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.075935 (rank : 46) | θ value | 0.00509761 (rank : 42) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.074182 (rank : 47) | θ value | 0.000270298 (rank : 39) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.072094 (rank : 48) | θ value | 0.0431538 (rank : 46) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.070952 (rank : 49) | θ value | 0.62314 (rank : 70) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.070698 (rank : 50) | θ value | 0.365318 (rank : 58) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.068796 (rank : 51) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.067754 (rank : 52) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.066001 (rank : 53) | θ value | 0.0252991 (rank : 45) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.065848 (rank : 54) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.065424 (rank : 55) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.065373 (rank : 56) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.060469 (rank : 57) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.060298 (rank : 58) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.060136 (rank : 59) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.059020 (rank : 60) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.056540 (rank : 61) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.056297 (rank : 62) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.056210 (rank : 63) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.056041 (rank : 64) | θ value | 0.279714 (rank : 57) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.055701 (rank : 65) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.053524 (rank : 66) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.052754 (rank : 67) | θ value | 0.0736092 (rank : 52) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.052341 (rank : 68) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.052337 (rank : 69) | θ value | 0.0961366 (rank : 53) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.052166 (rank : 70) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.050790 (rank : 71) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.049638 (rank : 72) | θ value | 0.0193708 (rank : 44) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.048987 (rank : 73) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.047698 (rank : 74) | θ value | 3.41135e-07 (rank : 37) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.047047 (rank : 75) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.047016 (rank : 76) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.044708 (rank : 77) | θ value | 0.365318 (rank : 59) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
RBM14_HUMAN
|
||||||
| NC score | 0.044434 (rank : 78) | θ value | 0.000602161 (rank : 40) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
RBM14_MOUSE
|
||||||
| NC score | 0.044394 (rank : 79) | θ value | 0.00390308 (rank : 41) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.044313 (rank : 80) | θ value | 0.0563607 (rank : 49) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.044180 (rank : 81) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.043158 (rank : 82) | θ value | 0.0431538 (rank : 47) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.042175 (rank : 83) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.041662 (rank : 84) | θ value | 3.77169e-06 (rank : 38) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.040752 (rank : 85) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.039844 (rank : 86) | θ value | 0.365318 (rank : 60) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.039721 (rank : 87) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.038197 (rank : 88) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.037505 (rank : 89) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.037180 (rank : 90) | θ value | 0.125558 (rank : 55) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.035372 (rank : 91) | θ value | 0.62314 (rank : 67) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.034996 (rank : 92) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.034769 (rank : 93) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.033852 (rank : 94) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.032830 (rank : 95) | θ value | 1.06291 (rank : 78) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.032216 (rank : 96) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.030532 (rank : 97) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.030510 (rank : 98) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CENPC_MOUSE
|
||||||
| NC score | 0.029754 (rank : 99) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49452 | Gene names | Cenpc1, Cenpc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.029212 (rank : 100) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PHAR2_HUMAN
|
||||||
| NC score | 0.028283 (rank : 101) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.027979 (rank : 102) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
NCOA5_MOUSE
|
||||||
| NC score | 0.027630 (rank : 103) | θ value | 0.813845 (rank : 74) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91W39 | Gene names | Ncoa5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
MUCEN_MOUSE
|
||||||
| NC score | 0.027581 (rank : 104) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.026109 (rank : 105) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.025649 (rank : 106) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.025241 (rank : 107) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.025059 (rank : 108) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.024403 (rank : 109) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.024185 (rank : 110) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.023449 (rank : 111) | θ value | 0.0563607 (rank : 51) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
CASL_HUMAN
|
||||||
| NC score | 0.022462 (rank : 112) | θ value | 0.279714 (rank : 56) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
ZCC2_HUMAN
|
||||||
| NC score | 0.019583 (rank : 113) | θ value | 0.62314 (rank : 71) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
NOL1_HUMAN
|
||||||
| NC score | 0.019399 (rank : 114) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
|
K1949_MOUSE
|
||||||
| NC score | 0.019320 (rank : 115) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
PAWR_HUMAN
|
||||||
| NC score | 0.019062 (rank : 116) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
YTHD1_HUMAN
|
||||||
| NC score | 0.018679 (rank : 117) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
TAB2_HUMAN
|
||||||
| NC score | 0.018208 (rank : 118) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYJ8, O94838, Q6I9W8, Q76N06, Q9UFP7 | Gene names | MAP3K7IP2, KIAA0733, TAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.017864 (rank : 119) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.017782 (rank : 120) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
TSYL1_MOUSE
|
||||||
| NC score | 0.017091 (rank : 121) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88852, Q3TKW0 | Gene names | Tspyl1, Tspyl | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific Y-encoded-like protein 1 (TSPY-like 1). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.016343 (rank : 122) | θ value | 1.06291 (rank : 77) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.016184 (rank : 123) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
DSRAD_MOUSE
|
||||||
| NC score | 0.015902 (rank : 124) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.014665 (rank : 125) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
YTHD1_MOUSE
|
||||||
| NC score | 0.013235 (rank : 126) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59326, Q3T9E2 | Gene names | Ythdf1 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1 homolog) (DACA-1 homolog). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.012043 (rank : 127) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.011563 (rank : 128) | θ value | 1.06291 (rank : 80) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.011519 (rank : 129) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.011017 (rank : 130) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.010927 (rank : 131) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.010419 (rank : 132) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
EMID2_HUMAN
|
||||||
| NC score | 0.010127 (rank : 133) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96A83 | Gene names | EMID2, COL26A1, EMU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
CNTN6_MOUSE
|
||||||
| NC score | 0.008935 (rank : 134) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JMB8, Q8C6X1 | Gene names | Cntn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (mNB-3). | |||||
|
CNTN6_HUMAN
|
||||||
| NC score | 0.008655 (rank : 135) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ52 | Gene names | CNTN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (hNB-3). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.008650 (rank : 136) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.008370 (rank : 137) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CNTN3_HUMAN
|
||||||
| NC score | 0.008044 (rank : 138) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P232, Q9H039 | Gene names | CNTN3, KIAA1496, PANG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
SCYL2_HUMAN
|
||||||
| NC score | 0.007611 (rank : 139) | θ value | 0.365318 (rank : 61) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 632 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3W7, Q96EF4, Q96ST4, Q9H7V5, Q9NVH3, Q9P2I7 | Gene names | SCYL2, CVAK104, KIAA1360 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
CNTN3_MOUSE
|
||||||
| NC score | 0.007284 (rank : 140) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07409 | Gene names | Cntn3, Pang, Pcs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
HXD12_MOUSE
|
||||||
| NC score | 0.007138 (rank : 141) | θ value | 0.0961366 (rank : 54) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23812 | Gene names | Hoxd12, Hox-4.7, Hoxd-12 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D12 (Hox-4.7) (Hox-5.6). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.006512 (rank : 142) | θ value | 0.62314 (rank : 69) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
CNTN4_MOUSE
|
||||||
| NC score | 0.006405 (rank : 143) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q69Z26, Q8BRT6, Q8CBD3 | Gene names | Cntn4, Kiaa3024 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.004534 (rank : 144) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
SHE_MOUSE
|
||||||
| NC score | 0.004038 (rank : 145) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BSD5, Q3TZT0 | Gene names | She | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein E. | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.003912 (rank : 146) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | 0.001718 (rank : 147) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
MIME_HUMAN
|
||||||
| NC score | 0.001295 (rank : 148) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20774, Q9UF90, Q9UNK5 | Gene names | OGN, OIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mimecan precursor (Osteoglycin) (Osteoinductive factor) (OIF). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.001203 (rank : 149) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MARK2_HUMAN
|
||||||
| NC score | -0.001815 (rank : 150) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | -0.003490 (rank : 151) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN579_HUMAN
|
||||||
| NC score | -0.003649 (rank : 152) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NAF0 | Gene names | ZNF579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 579. | |||||
|
ERBB2_MOUSE
|
||||||
| NC score | -0.004267 (rank : 153) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||