Please be patient as the page loads
|
NOL1_HUMAN
|
||||||
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NOL1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 2) | NC score | 0.085095 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 3) | NC score | 0.082343 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.063627 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.279714 (rank : 5) | NC score | 0.067189 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RRMJ1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 6) | NC score | 0.081495 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UET6, O75670 | Gene names | FTSJ1 | |||
|
Domain Architecture |
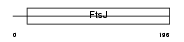
|
|||||
| Description | Putative ribosomal RNA methyltransferase 1 (EC 2.1.1.-) (rRNA (uridine-2'-O-)-methyltransferase). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.085167 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
SHAN2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.044652 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.013604 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
SASH1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.047749 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
AMPD2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.052042 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01433, Q14856, Q14857, Q16686, Q16687, Q16688, Q16729, Q96IA1, Q9UDX8, Q9UDX9, Q9UMU4 | Gene names | AMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase isoform L). | |||||
|
AMPD2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.052016 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBT5, Q91YI2 | Gene names | Ampd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6). | |||||
|
CD008_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.064069 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.022699 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.055889 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
SEBP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.055230 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
ANFY1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.026242 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANFY1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.026145 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ISL2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.016358 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CXV0 | Gene names | Isl2 | |||
|
Domain Architecture |
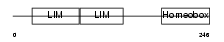
|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.017930 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.047983 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.010762 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.037547 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.040527 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.013714 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.014943 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.044436 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
CE005_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.022981 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.037971 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.037942 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
HMGA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.071208 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52926 | Gene names | HMGA2, HMGIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMGI-C (High mobility group AT-hook protein 2). | |||||
|
MAGD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.019399 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.054578 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PKHG4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.028079 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.044366 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.058011 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.028620 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.021794 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.021980 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.023121 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANXA7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.012615 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07076 | Gene names | Anxa7, Anx7 | |||
|
Domain Architecture |
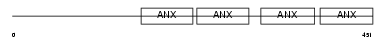
|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.012563 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.035498 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.051618 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.025278 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SFRS8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.047547 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.048590 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
ACSA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.017930 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXG4 | Gene names | Acss2, Acas2, Acecs1 | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-coenzyme A synthetase, cytoplasmic (EC 6.2.1.1) (Acetate--CoA ligase) (Acyl-activating enzyme) (Acetyl-CoA synthetase) (ACS) (AceCS) (Acyl-CoA synthetase short-chain family member 2). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.013137 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
DDX18_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.007673 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.001900 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.015153 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
HXC10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.007558 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYD6, O15219, O15220, Q9BVD5 | Gene names | HOXC10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C10. | |||||
|
IKBA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.014660 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1E3, Q80ZX5 | Gene names | Nfkbia, Ikba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B inhibitor alpha (I-kappa-B-alpha) (IkappaBalpha) (IKB- alpha). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.028639 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.023202 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.036272 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PSIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.035131 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.028956 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.019324 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ZHX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.017966 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
AKAP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.021474 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
GASP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.014751 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.030459 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
KCTD3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.014358 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BFX3, Q6P7X4 | Gene names | Kctd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.029793 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.015878 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PCD16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.004061 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
SOX11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.010423 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SYMPK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.019838 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
TOP2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.017356 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01320 | Gene names | Top2a, Top-2, Top2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
UB7I1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.034271 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.033702 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
ZFY19_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.023970 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
NOL1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.085167 (rank : 2) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.085095 (rank : 3) | θ value | 0.0330416 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.082343 (rank : 4) | θ value | 0.0961366 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
RRMJ1_HUMAN
|
||||||
| NC score | 0.081495 (rank : 5) | θ value | 0.365318 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UET6, O75670 | Gene names | FTSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ribosomal RNA methyltransferase 1 (EC 2.1.1.-) (rRNA (uridine-2'-O-)-methyltransferase). | |||||
|
HMGA2_HUMAN
|
||||||
| NC score | 0.071208 (rank : 6) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52926 | Gene names | HMGA2, HMGIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMGI-C (High mobility group AT-hook protein 2). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.067189 (rank : 7) | θ value | 0.279714 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CD008_HUMAN
|
||||||
| NC score | 0.064069 (rank : 8) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.063627 (rank : 9) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.058011 (rank : 10) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.055889 (rank : 11) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
SEBP2_HUMAN
|
||||||
| NC score | 0.055230 (rank : 12) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.054578 (rank : 13) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
AMPD2_HUMAN
|
||||||
| NC score | 0.052042 (rank : 14) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01433, Q14856, Q14857, Q16686, Q16687, Q16688, Q16729, Q96IA1, Q9UDX8, Q9UDX9, Q9UMU4 | Gene names | AMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase isoform L). | |||||
|
AMPD2_MOUSE
|
||||||
| NC score | 0.052016 (rank : 15) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBT5, Q91YI2 | Gene names | Ampd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6). | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.051618 (rank : 16) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.048590 (rank : 17) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.047983 (rank : 18) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
SASH1_MOUSE
|
||||||
| NC score | 0.047749 (rank : 19) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
SFRS8_HUMAN
|
||||||
| NC score | 0.047547 (rank : 20) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
SHAN2_MOUSE
|
||||||
| NC score | 0.044652 (rank : 21) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.044436 (rank : 22) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.044366 (rank : 23) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.040527 (rank : 24) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.037971 (rank : 25) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.037942 (rank : 26) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.037547 (rank : 27) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.036272 (rank : 28) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.035498 (rank : 29) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
PSIP1_HUMAN
|
||||||
| NC score | 0.035131 (rank : 30) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
UB7I1_HUMAN
|
||||||
| NC score | 0.034271 (rank : 31) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.033702 (rank : 32) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.030459 (rank : 33) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.029793 (rank : 34) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.028956 (rank : 35) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.028639 (rank : 36) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.028620 (rank : 37) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PKHG4_HUMAN
|
||||||
| NC score | 0.028079 (rank : 38) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
ANFY1_HUMAN
|
||||||
| NC score | 0.026242 (rank : 39) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANFY1_MOUSE
|
||||||
| NC score | 0.026145 (rank : 40) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.025278 (rank : 41) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
ZFY19_HUMAN
|
||||||
| NC score | 0.023970 (rank : 42) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.023202 (rank : 43) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.023121 (rank : 44) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.022981 (rank : 45) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.022699 (rank : 46) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.021980 (rank : 47) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.021794 (rank : 48) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
AKAP6_HUMAN
|
||||||
| NC score | 0.021474 (rank : 49) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
SYMPK_HUMAN
|
||||||
| NC score | 0.019838 (rank : 50) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
MAGD1_MOUSE
|
||||||
| NC score | 0.019399 (rank : 51) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.019324 (rank : 52) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ZHX2_MOUSE
|
||||||
| NC score | 0.017966 (rank : 53) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.017930 (rank : 54) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
ACSA_MOUSE
|
||||||
| NC score | 0.017930 (rank : 55) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXG4 | Gene names | Acss2, Acas2, Acecs1 | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-coenzyme A synthetase, cytoplasmic (EC 6.2.1.1) (Acetate--CoA ligase) (Acyl-activating enzyme) (Acetyl-CoA synthetase) (ACS) (AceCS) (Acyl-CoA synthetase short-chain family member 2). | |||||
|
TOP2A_MOUSE
|
||||||
| NC score | 0.017356 (rank : 56) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01320 | Gene names | Top2a, Top-2, Top2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
ISL2_MOUSE
|
||||||
| NC score | 0.016358 (rank : 57) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CXV0 | Gene names | Isl2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.015878 (rank : 58) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.015153 (rank : 59) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.014943 (rank : 60) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
GASP1_HUMAN
|
||||||
| NC score | 0.014751 (rank : 61) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
IKBA_MOUSE
|
||||||
| NC score | 0.014660 (rank : 62) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1E3, Q80ZX5 | Gene names | Nfkbia, Ikba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B inhibitor alpha (I-kappa-B-alpha) (IkappaBalpha) (IKB- alpha). | |||||
|
KCTD3_MOUSE
|
||||||
| NC score | 0.014358 (rank : 63) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BFX3, Q6P7X4 | Gene names | Kctd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3. | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.013714 (rank : 64) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.013604 (rank : 65) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.013137 (rank : 66) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
ANXA7_MOUSE
|
||||||
| NC score | 0.012615 (rank : 67) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07076 | Gene names | Anxa7, Anx7 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.012563 (rank : 68) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.010762 (rank : 69) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
SOX11_MOUSE
|
||||||
| NC score | 0.010423 (rank : 70) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.007673 (rank : 71) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
HXC10_HUMAN
|
||||||
| NC score | 0.007558 (rank : 72) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYD6, O15219, O15220, Q9BVD5 | Gene names | HOXC10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C10. | |||||
|
PCD16_HUMAN
|
||||||
| NC score | 0.004061 (rank : 73) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
EVI1_HUMAN
|
||||||
| NC score | 0.001900 (rank : 74) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||