Please be patient as the page loads
|
AMPD2_HUMAN
|
||||||
| SwissProt Accessions | Q01433, Q14856, Q14857, Q16686, Q16687, Q16688, Q16729, Q96IA1, Q9UDX8, Q9UDX9, Q9UMU4 | Gene names | AMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase isoform L). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AMPD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.983364 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23109 | Gene names | AMPD1 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 1 (EC 3.5.4.6) (Myoadenylate deaminase) (AMP deaminase isoform M). | |||||
|
AMPD2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01433, Q14856, Q14857, Q16686, Q16687, Q16688, Q16729, Q96IA1, Q9UDX8, Q9UDX9, Q9UMU4 | Gene names | AMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase isoform L). | |||||
|
AMPD2_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.998819 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DBT5, Q91YI2 | Gene names | Ampd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6). | |||||
|
AMPD3_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.979338 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01432 | Gene names | AMPD3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (Erythrocyte AMP deaminase). | |||||
|
AMPD3_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.979804 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08739, O88692 | Gene names | Ampd3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (AMP deaminase H-type) (Heart-type AMPD). | |||||
|
CECR1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.233273 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZK5, Q96K41 | Gene names | CECR1 | |||
|
Domain Architecture |
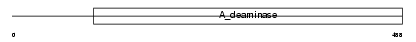
|
|||||
| Description | Cat eye syndrome critical region protein 1 precursor. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.017021 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.025764 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
NOL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.052042 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
|
NBEA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.024343 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
ZN406_HUMAN
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.001970 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P243, Q3MIM5, Q6PJ01, Q75PJ7, Q75PJ9, Q86X64 | Gene names | ZNF406, KIAA1485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406 (Protein ZFAT). | |||||
|
CING_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.007754 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.008160 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
NBEA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.022331 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFP9, Q9HCM8, Q9NSU1, Q9NW98, Q9Y6J1 | Gene names | NBEA, BCL8B, KIAA1544, LYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2) (Protein BCL8B). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.004604 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DPOG2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.028350 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZM2, O35614 | Gene names | Polg2, Mtpolb | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase subunit gamma 2, mitochondrial precursor (EC 2.7.7.7) (Mitochondrial DNA polymerase accessory subunit) (PolG-beta) (MtPolB) (DNA polymerase gamma accessory 55 kDa subunit) (p55). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.012804 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
IKKB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.003572 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88351, Q9R1J6 | Gene names | Ikbkb, Ikkb | |||
|
Domain Architecture |
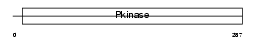
|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.014937 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.001125 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
CEP55_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.012627 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q53EZ4, Q32WF5, Q3MV20, Q5VY28, Q6N034, Q96H32, Q9NVS7 | Gene names | CEP55, C10orf3, URCC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55 (Up-regulated in colon cancer 6). | |||||
|
CO9A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.003744 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
CP135_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.004281 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
PAR10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.017520 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.003753 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
AMPD2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01433, Q14856, Q14857, Q16686, Q16687, Q16688, Q16729, Q96IA1, Q9UDX8, Q9UDX9, Q9UMU4 | Gene names | AMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase isoform L). | |||||
|
AMPD2_MOUSE
|
||||||
| NC score | 0.998819 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DBT5, Q91YI2 | Gene names | Ampd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6). | |||||
|
AMPD1_HUMAN
|
||||||
| NC score | 0.983364 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23109 | Gene names | AMPD1 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 1 (EC 3.5.4.6) (Myoadenylate deaminase) (AMP deaminase isoform M). | |||||
|
AMPD3_MOUSE
|
||||||
| NC score | 0.979804 (rank : 4) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08739, O88692 | Gene names | Ampd3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (AMP deaminase H-type) (Heart-type AMPD). | |||||
|
AMPD3_HUMAN
|
||||||
| NC score | 0.979338 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01432 | Gene names | AMPD3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (Erythrocyte AMP deaminase). | |||||
|
CECR1_HUMAN
|
||||||
| NC score | 0.233273 (rank : 6) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZK5, Q96K41 | Gene names | CECR1 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 1 precursor. | |||||
|
NOL1_HUMAN
|
||||||
| NC score | 0.052042 (rank : 7) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
|
DPOG2_MOUSE
|
||||||
| NC score | 0.028350 (rank : 8) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZM2, O35614 | Gene names | Polg2, Mtpolb | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase subunit gamma 2, mitochondrial precursor (EC 2.7.7.7) (Mitochondrial DNA polymerase accessory subunit) (PolG-beta) (MtPolB) (DNA polymerase gamma accessory 55 kDa subunit) (p55). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.025764 (rank : 9) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
NBEA_MOUSE
|
||||||
| NC score | 0.024343 (rank : 10) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
NBEA_HUMAN
|
||||||
| NC score | 0.022331 (rank : 11) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFP9, Q9HCM8, Q9NSU1, Q9NW98, Q9Y6J1 | Gene names | NBEA, BCL8B, KIAA1544, LYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2) (Protein BCL8B). | |||||
|
PAR10_HUMAN
|
||||||
| NC score | 0.017520 (rank : 12) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.017021 (rank : 13) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.014937 (rank : 14) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.012804 (rank : 15) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
CEP55_HUMAN
|
||||||
| NC score | 0.012627 (rank : 16) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q53EZ4, Q32WF5, Q3MV20, Q5VY28, Q6N034, Q96H32, Q9NVS7 | Gene names | CEP55, C10orf3, URCC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55 (Up-regulated in colon cancer 6). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.008160 (rank : 17) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.007754 (rank : 18) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.004604 (rank : 19) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.004281 (rank : 20) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.003753 (rank : 21) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
CO9A2_MOUSE
|
||||||
| NC score | 0.003744 (rank : 22) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
IKKB_MOUSE
|
||||||
| NC score | 0.003572 (rank : 23) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88351, Q9R1J6 | Gene names | Ikbkb, Ikkb | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
ZN406_HUMAN
|
||||||
| NC score | 0.001970 (rank : 24) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P243, Q3MIM5, Q6PJ01, Q75PJ7, Q75PJ9, Q86X64 | Gene names | ZNF406, KIAA1485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406 (Protein ZFAT). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.001125 (rank : 25) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||