Please be patient as the page loads
|
AMPD3_MOUSE
|
||||||
| SwissProt Accessions | O08739, O88692 | Gene names | Ampd3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (AMP deaminase H-type) (Heart-type AMPD). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AMPD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991316 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23109 | Gene names | AMPD1 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 1 (EC 3.5.4.6) (Myoadenylate deaminase) (AMP deaminase isoform M). | |||||
|
AMPD2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.979804 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01433, Q14856, Q14857, Q16686, Q16687, Q16688, Q16729, Q96IA1, Q9UDX8, Q9UDX9, Q9UMU4 | Gene names | AMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase isoform L). | |||||
|
AMPD2_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.981264 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBT5, Q91YI2 | Gene names | Ampd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6). | |||||
|
AMPD3_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.997550 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01432 | Gene names | AMPD3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (Erythrocyte AMP deaminase). | |||||
|
AMPD3_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08739, O88692 | Gene names | Ampd3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (AMP deaminase H-type) (Heart-type AMPD). | |||||
|
CECR1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 6) | NC score | 0.268619 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZK5, Q96K41 | Gene names | CECR1 | |||
|
Domain Architecture |
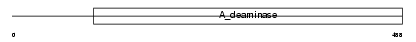
|
|||||
| Description | Cat eye syndrome critical region protein 1 precursor. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 7) | NC score | 0.024505 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.079513 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
NELFA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.073992 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
NOP14_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.025621 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.006481 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
PDGFD_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.025733 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.018052 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
AHTF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.017753 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
CD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.021827 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |
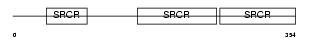
|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
RRMJ3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.025312 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
ABR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.012614 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
ORC3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.026157 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |
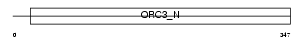
|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
CAD23_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.007586 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PF4, Q99NH1, Q9D4N9 | Gene names | Cdh23 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.009938 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.005819 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.010669 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
K0090_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.013755 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C7X2, Q3TCG4, Q6ZQJ4, Q8K3W8 | Gene names | Kiaa0090 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0090 precursor. | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.011691 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.007023 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CAD23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.006608 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H251, Q6UWW1, Q9H4K9 | Gene names | CDH23, KIAA1774 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
CD6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.012451 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |
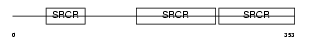
|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
TECT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.010991 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ64, Q3UMP0, Q8BUE2 | Gene names | Tect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-1 precursor. | |||||
|
U2AFL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.008590 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.001828 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
FA8A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.015150 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBU6 | Gene names | FAM8A1, AHCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM8A1 (Autosomal highly conserved protein). | |||||
|
FBW1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.002178 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y297, Q9Y213 | Gene names | BTRC, BTRCP, FBW1A, FBXW1A | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 1A (F-box and WD repeats protein beta-TrCP) (E3RSIkappaB) (pIkappaBalpha-E3 receptor subunit). | |||||
|
INVO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.004576 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.007562 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
AMPD3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08739, O88692 | Gene names | Ampd3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (AMP deaminase H-type) (Heart-type AMPD). | |||||
|
AMPD3_HUMAN
|
||||||
| NC score | 0.997550 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01432 | Gene names | AMPD3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (Erythrocyte AMP deaminase). | |||||
|
AMPD1_HUMAN
|
||||||
| NC score | 0.991316 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23109 | Gene names | AMPD1 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 1 (EC 3.5.4.6) (Myoadenylate deaminase) (AMP deaminase isoform M). | |||||
|
AMPD2_MOUSE
|
||||||
| NC score | 0.981264 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBT5, Q91YI2 | Gene names | Ampd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6). | |||||
|
AMPD2_HUMAN
|
||||||
| NC score | 0.979804 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01433, Q14856, Q14857, Q16686, Q16687, Q16688, Q16729, Q96IA1, Q9UDX8, Q9UDX9, Q9UMU4 | Gene names | AMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase isoform L). | |||||
|
CECR1_HUMAN
|
||||||
| NC score | 0.268619 (rank : 6) | θ value | 0.00020696 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZK5, Q96K41 | Gene names | CECR1 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 1 precursor. | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.079513 (rank : 7) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
NELFA_HUMAN
|
||||||
| NC score | 0.073992 (rank : 8) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
ORC3_MOUSE
|
||||||
| NC score | 0.026157 (rank : 9) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
PDGFD_MOUSE
|
||||||
| NC score | 0.025733 (rank : 10) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.025621 (rank : 11) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
RRMJ3_MOUSE
|
||||||
| NC score | 0.025312 (rank : 12) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.024505 (rank : 13) | θ value | 0.0961366 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CD6_HUMAN
|
||||||
| NC score | 0.021827 (rank : 14) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.018052 (rank : 15) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
AHTF1_HUMAN
|
||||||
| NC score | 0.017753 (rank : 16) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
FA8A1_HUMAN
|
||||||
| NC score | 0.015150 (rank : 17) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBU6 | Gene names | FAM8A1, AHCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM8A1 (Autosomal highly conserved protein). | |||||
|
K0090_MOUSE
|
||||||
| NC score | 0.013755 (rank : 18) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C7X2, Q3TCG4, Q6ZQJ4, Q8K3W8 | Gene names | Kiaa0090 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0090 precursor. | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.012614 (rank : 19) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
CD6_MOUSE
|
||||||
| NC score | 0.012451 (rank : 20) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.011691 (rank : 21) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TECT1_MOUSE
|
||||||
| NC score | 0.010991 (rank : 22) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ64, Q3UMP0, Q8BUE2 | Gene names | Tect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-1 precursor. | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.010669 (rank : 23) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.009938 (rank : 24) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
U2AFL_MOUSE
|
||||||
| NC score | 0.008590 (rank : 25) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
CAD23_MOUSE
|
||||||
| NC score | 0.007586 (rank : 26) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PF4, Q99NH1, Q9D4N9 | Gene names | Cdh23 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.007562 (rank : 27) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.007023 (rank : 28) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CAD23_HUMAN
|
||||||
| NC score | 0.006608 (rank : 29) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H251, Q6UWW1, Q9H4K9 | Gene names | CDH23, KIAA1774 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.006481 (rank : 30) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.005819 (rank : 31) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.004576 (rank : 32) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
FBW1A_HUMAN
|
||||||
| NC score | 0.002178 (rank : 33) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y297, Q9Y213 | Gene names | BTRC, BTRCP, FBW1A, FBXW1A | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 1A (F-box and WD repeats protein beta-TrCP) (E3RSIkappaB) (pIkappaBalpha-E3 receptor subunit). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.001828 (rank : 34) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||