Please be patient as the page loads
|
CD6_HUMAN
|
||||||
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CD6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
CD6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992618 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
DMBT1_HUMAN
|
||||||
| θ value | 6.53529e-67 (rank : 3) | NC score | 0.744530 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | 7.24236e-58 (rank : 4) | NC score | 0.675898 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
C163A_MOUSE
|
||||||
| θ value | 1.01294e-51 (rank : 5) | NC score | 0.912961 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163A_HUMAN
|
||||||
| θ value | 2.25659e-51 (rank : 6) | NC score | 0.911338 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163B_HUMAN
|
||||||
| θ value | 2.25659e-51 (rank : 7) | NC score | 0.909177 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
CD5L_HUMAN
|
||||||
| θ value | 6.14528e-49 (rank : 8) | NC score | 0.936369 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
|
NETR_HUMAN
|
||||||
| θ value | 1.12253e-42 (rank : 9) | NC score | 0.437952 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
CD5L_MOUSE
|
||||||
| θ value | 3.61106e-41 (rank : 10) | NC score | 0.931126 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
NETR_MOUSE
|
||||||
| θ value | 1.32909e-35 (rank : 11) | NC score | 0.423049 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
LOXL3_MOUSE
|
||||||
| θ value | 4.27553e-34 (rank : 12) | NC score | 0.804399 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
LOXL2_MOUSE
|
||||||
| θ value | 7.29293e-34 (rank : 13) | NC score | 0.806213 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
LOXL3_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 14) | NC score | 0.801097 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
LOXL4_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 15) | NC score | 0.802600 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL2_HUMAN
|
||||||
| θ value | 9.8567e-31 (rank : 16) | NC score | 0.802122 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
LOXL4_MOUSE
|
||||||
| θ value | 1.4233e-29 (rank : 17) | NC score | 0.801757 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
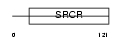
LG3BP_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 18) | NC score | 0.840434 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
|
MSRE_MOUSE
|
||||||
| θ value | 4.29542e-18 (rank : 19) | NC score | 0.656788 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
MSRE_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 20) | NC score | 0.598152 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P21757, P21759, Q45F10 | Gene names | MSR1, SCARA1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor class A member 1) (CD204 antigen). | |||||
|
MARCO_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 21) | NC score | 0.411170 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
MARCO_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 22) | NC score | 0.508574 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UEW3, Q9Y5S3 | Gene names | MARCO, SCARA2 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure) (Scavenger receptor class A member 2). | |||||
|
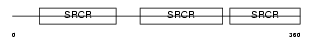
CD5_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 23) | NC score | 0.622636 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
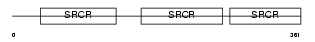
CD5_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 24) | NC score | 0.581679 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
ENTK_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 25) | NC score | 0.116889 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
ENTK_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 26) | NC score | 0.103410 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
SYNP2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.036600 (rank : 89) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 28) | NC score | 0.013648 (rank : 98) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CFAI_MOUSE
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.111972 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
HSH2D_MOUSE
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.026346 (rank : 91) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
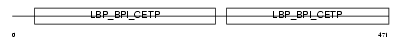
CETP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.029323 (rank : 90) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11597, Q13987, Q13988 | Gene names | CETP | |||
|
Domain Architecture |
|
|||||
| Description | Cholesteryl ester transfer protein precursor (Lipid transfer protein I). | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.076885 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
DMRTD_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.021017 (rank : 94) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.011596 (rank : 100) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
ADA12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.009652 (rank : 104) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43184, O60470 | Gene names | ADAM12, MLTN | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.022343 (rank : 92) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.008371 (rank : 107) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
AMPD3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.018481 (rank : 95) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01432 | Gene names | AMPD3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (Erythrocyte AMP deaminase). | |||||
|
AMPD3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.021827 (rank : 93) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08739, O88692 | Gene names | Ampd3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (AMP deaminase H-type) (Heart-type AMPD). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.004296 (rank : 113) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.010636 (rank : 102) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CENPC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.015274 (rank : 97) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.003109 (rank : 115) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.008272 (rank : 108) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SON_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.017595 (rank : 96) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
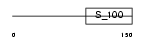
EFHD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.010519 (rank : 103) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D4J1, Q543U4 | Gene names | Efhd1, Sws2 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
GRN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.009338 (rank : 105) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.010637 (rank : 101) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.005402 (rank : 112) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.000626 (rank : 117) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.009120 (rank : 106) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NUPL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.013646 (rank : 99) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
HXC10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.001819 (rank : 116) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYD6, O15219, O15220, Q9BVD5 | Gene names | HOXC10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C10. | |||||
|
MARK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | -0.003366 (rank : 120) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96L34, Q8NG37, Q96JG7, Q96SQ2, Q9BYD8 | Gene names | MARK4, KIAA1860, MARKL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 4 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase-like 1). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | -0.002405 (rank : 118) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TMPS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.058686 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
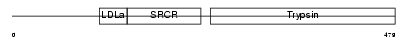
TMPS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.049233 (rank : 88) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
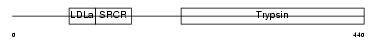
TMPS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.079026 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
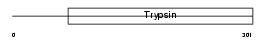
TMPS5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.059719 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ER04, Q9ER02, Q9ER03 | Gene names | Tmprss5 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
ZN366_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | -0.003206 (rank : 119) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N895, Q7RTV4 | Gene names | ZNF366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 366. | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.003315 (rank : 114) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.007420 (rank : 109) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
PAX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.006855 (rank : 111) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32114 | Gene names | Pax2, Pax-2 | |||
|
Domain Architecture |
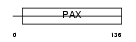
|
|||||
| Description | Paired box protein Pax-2. | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.006906 (rank : 110) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
ZN606_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | -0.003670 (rank : 121) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXB4, Q8NE04, Q96JH5 | Gene names | ZNF606, KIAA1852, ZNF328 | |||
|
Domain Architecture |
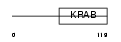
|
|||||
| Description | Zinc finger protein 606 (Zinc finger protein 328). | |||||
|
BMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.054873 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.055147 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
C1RA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.052508 (rank : 82) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
C1RB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.053321 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
C1R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.053948 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
C1S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.056322 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
CFAI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.108825 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CORIN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.058230 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
CS1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.056428 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
CS1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.057244 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
CUZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.112888 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.116962 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
KREM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.051867 (rank : 84) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |
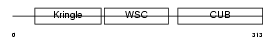
|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
LOXL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.267108 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
LOXL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.266159 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
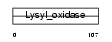
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
LRP12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.053113 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
LRP12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.053213 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LYOX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.280600 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28300, Q5FWF0 | Gene names | LOX | |||
|
Domain Architecture |
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase). | |||||
|
LYOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.278459 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
MASP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.057179 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.056448 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
MASP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.055570 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MASP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.052893 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MFRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.052553 (rank : 81) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.053418 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
NETO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.052193 (rank : 83) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.051669 (rank : 85) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.053901 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.054125 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.056014 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PCOC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.056853 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.062700 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.063634 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
ST14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.061045 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
ST14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.062864 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
TECTB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.054123 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PL2 | Gene names | TECTB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
TECTB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.054154 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08524 | Gene names | Tectb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
TGBR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.054441 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03167, Q5T2T4, Q5U731, Q9UGI2 | Gene names | TGFBR3 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
TGBR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053027 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88393, Q6NS72 | Gene names | Tgfbr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
TLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.058971 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
TLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.059921 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
TLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.057251 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.058006 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TMPS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.081672 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
TMPS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.050309 (rank : 87) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRS4, Q5XKQ6, Q6UX37, Q9NZA5 | Gene names | TMPRSS4, TMPRSS3 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Membrane-type serine protease 2) (MT-SP2). | |||||
|
TMPS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.060593 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCA5 | Gene names | Tmprss4, Cap2 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Channel-activating protease 2) (mCAP2). | |||||
|
TMPS5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.052839 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H3S3 | Gene names | TMPRSS5 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
TMPS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.053992 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TMPS7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.051470 (rank : 86) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BIK6 | Gene names | Tmprss7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.057142 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TSG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.059809 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.059791 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
ZP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.063075 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05996 | Gene names | ZP2, ZPA | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
ZP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.057638 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20239 | Gene names | Zp2, Zp-2, Zpa | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
ZP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.052726 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10761 | Gene names | Zp3, Zp-3, Zpc | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 precursor (Zona pellucida glycoprotein ZP3) (Sperm receptor) (Zona pellucida protein C). | |||||
|
ZP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.056013 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12836 | Gene names | ZP4, ZPB | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 4 precursor (Zona pellucida sperm-binding protein B). | |||||
|
CD6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
CD6_MOUSE
|
||||||
| NC score | 0.992618 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
CD5L_HUMAN
|
||||||
| NC score | 0.936369 (rank : 3) | θ value | 6.14528e-49 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
|
CD5L_MOUSE
|
||||||
| NC score | 0.931126 (rank : 4) | θ value | 3.61106e-41 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
C163A_MOUSE
|
||||||
| NC score | 0.912961 (rank : 5) | θ value | 1.01294e-51 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163A_HUMAN
|
||||||
| NC score | 0.911338 (rank : 6) | θ value | 2.25659e-51 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163B_HUMAN
|
||||||
| NC score | 0.909177 (rank : 7) | θ value | 2.25659e-51 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
LG3BP_HUMAN
|
||||||
| NC score | 0.840434 (rank : 8) | θ value | 3.75424e-22 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
|
LOXL2_MOUSE
|
||||||
| NC score | 0.806213 (rank : 9) | θ value | 7.29293e-34 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
LOXL3_MOUSE
|
||||||
| NC score | 0.804399 (rank : 10) | θ value | 4.27553e-34 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
LOXL4_HUMAN
|
||||||
| NC score | 0.802600 (rank : 11) | θ value | 8.91499e-32 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL2_HUMAN
|
||||||
| NC score | 0.802122 (rank : 12) | θ value | 9.8567e-31 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
LOXL4_MOUSE
|
||||||
| NC score | 0.801757 (rank : 13) | θ value | 1.4233e-29 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL3_HUMAN
|
||||||
| NC score | 0.801097 (rank : 14) | θ value | 8.91499e-32 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
DMBT1_HUMAN
|
||||||
| NC score | 0.744530 (rank : 15) | θ value | 6.53529e-67 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.675898 (rank : 16) | θ value | 7.24236e-58 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
MSRE_MOUSE
|
||||||
| NC score | 0.656788 (rank : 17) | θ value | 4.29542e-18 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
CD5_MOUSE
|
||||||
| NC score | 0.622636 (rank : 18) | θ value | 4.59992e-12 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
MSRE_HUMAN
|
||||||
| NC score | 0.598152 (rank : 19) | θ value | 2.7842e-17 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P21757, P21759, Q45F10 | Gene names | MSR1, SCARA1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor class A member 1) (CD204 antigen). | |||||
|
CD5_HUMAN
|
||||||
| NC score | 0.581679 (rank : 20) | θ value | 1.38499e-08 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
MARCO_HUMAN
|
||||||
| NC score | 0.508574 (rank : 21) | θ value | 8.38298e-14 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UEW3, Q9Y5S3 | Gene names | MARCO, SCARA2 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure) (Scavenger receptor class A member 2). | |||||
|
NETR_HUMAN
|
||||||
| NC score | 0.437952 (rank : 22) | θ value | 1.12253e-42 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
NETR_MOUSE
|
||||||
| NC score | 0.423049 (rank : 23) | θ value | 1.32909e-35 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
MARCO_MOUSE
|
||||||
| NC score | 0.411170 (rank : 24) | θ value | 8.10077e-17 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
LYOX_HUMAN
|
||||||
| NC score | 0.280600 (rank : 25) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28300, Q5FWF0 | Gene names | LOX | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase). | |||||
|
LYOX_MOUSE
|
||||||
| NC score | 0.278459 (rank : 26) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
LOXL1_HUMAN
|
||||||
| NC score | 0.267108 (rank : 27) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
LOXL1_MOUSE
|
||||||
| NC score | 0.266159 (rank : 28) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.116962 (rank : 29) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
ENTK_MOUSE
|
||||||
| NC score | 0.116889 (rank : 30) | θ value | 0.000121331 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
CUZD1_HUMAN
|
||||||
| NC score | 0.112888 (rank : 31) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CFAI_MOUSE
|
||||||
| NC score | 0.111972 (rank : 32) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CFAI_HUMAN
|
||||||
| NC score | 0.108825 (rank : 33) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
ENTK_HUMAN
|
||||||
| NC score | 0.103410 (rank : 34) | θ value | 0.00228821 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
TMPS3_HUMAN
|
||||||
| NC score | 0.081672 (rank : 35) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
TMPS3_MOUSE
|
||||||
| NC score | 0.079026 (rank : 36) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.076885 (rank : 37) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
PCOC2_MOUSE
|
||||||
| NC score | 0.063634 (rank : 38) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
ZP2_HUMAN
|
||||||
| NC score | 0.063075 (rank : 39) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05996 | Gene names | ZP2, ZPA | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
ST14_MOUSE
|
||||||
| NC score | 0.062864 (rank : 40) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
PCOC2_HUMAN
|
||||||
| NC score | 0.062700 (rank : 41) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
ST14_HUMAN
|
||||||
| NC score | 0.061045 (rank : 42) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
TMPS4_MOUSE
|
||||||
| NC score | 0.060593 (rank : 43) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCA5 | Gene names | Tmprss4, Cap2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Channel-activating protease 2) (mCAP2). | |||||
|
TLL1_MOUSE
|
||||||
| NC score | 0.059921 (rank : 44) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.059809 (rank : 45) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.059791 (rank : 46) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
TMPS5_MOUSE
|
||||||
| NC score | 0.059719 (rank : 47) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ER04, Q9ER02, Q9ER03 | Gene names | Tmprss5 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
TLL1_HUMAN
|
||||||
| NC score | 0.058971 (rank : 48) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
TMPS2_HUMAN
|
||||||
| NC score | 0.058686 (rank : 49) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.058230 (rank : 50) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
TLL2_MOUSE
|
||||||
| NC score | 0.058006 (rank : 51) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
ZP2_MOUSE
|
||||||
| NC score | 0.057638 (rank : 52) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20239 | Gene names | Zp2, Zp-2, Zpa | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
TLL2_HUMAN
|
||||||
| NC score | 0.057251 (rank : 53) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
CS1B_MOUSE
|
||||||
| NC score | 0.057244 (rank : 54) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
MASP1_HUMAN
|
||||||
| NC score | 0.057179 (rank : 55) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.057142 (rank : 56) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
PCOC1_MOUSE
|
||||||
| NC score | 0.056853 (rank : 57) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
MASP1_MOUSE
|
||||||
| NC score | 0.056448 (rank : 58) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
CS1A_MOUSE
|
||||||
| NC score | 0.056428 (rank : 59) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
C1S_HUMAN
|
||||||
| NC score | 0.056322 (rank : 60) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.056014 (rank : 61) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
ZP4_HUMAN
|
||||||
| NC score | 0.056013 (rank : 62) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12836 | Gene names | ZP4, ZPB | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 4 precursor (Zona pellucida sperm-binding protein B). | |||||
|
MASP2_HUMAN
|
||||||
| NC score | 0.055570 (rank : 63) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
BMP1_MOUSE
|
||||||
| NC score | 0.055147 (rank : 64) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_HUMAN
|
||||||
| NC score | 0.054873 (rank : 65) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
TGBR3_HUMAN
|
||||||
| NC score | 0.054441 (rank : 66) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03167, Q5T2T4, Q5U731, Q9UGI2 | Gene names | TGFBR3 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
TECTB_MOUSE
|
||||||
| NC score | 0.054154 (rank : 67) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08524 | Gene names | Tectb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
NETO2_MOUSE
|
||||||
| NC score | 0.054125 (rank : 68) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
TECTB_HUMAN
|
||||||
| NC score | 0.054123 (rank : 69) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PL2 | Gene names | TECTB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
TMPS7_HUMAN
|
||||||
| NC score | 0.053992 (rank : 70) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
C1R_HUMAN
|
||||||
| NC score | 0.053948 (rank : 71) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
NETO2_HUMAN
|
||||||
| NC score | 0.053901 (rank : 72) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.053418 (rank : 73) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
C1RB_MOUSE
|
||||||
| NC score | 0.053321 (rank : 74) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
LRP12_MOUSE
|
||||||
| NC score | 0.053213 (rank : 75) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LRP12_HUMAN
|
||||||
| NC score | 0.053113 (rank : 76) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
TGBR3_MOUSE
|
||||||
| NC score | 0.053027 (rank : 77) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88393, Q6NS72 | Gene names | Tgfbr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
MASP2_MOUSE
|
||||||
| NC score | 0.052893 (rank : 78) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
TMPS5_HUMAN
|
||||||
| NC score | 0.052839 (rank : 79) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H3S3 | Gene names | TMPRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
ZP3_MOUSE
|
||||||
| NC score | 0.052726 (rank : 80) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10761 | Gene names | Zp3, Zp-3, Zpc | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 precursor (Zona pellucida glycoprotein ZP3) (Sperm receptor) (Zona pellucida protein C). | |||||
|
MFRP_HUMAN
|
||||||
| NC score | 0.052553 (rank : 81) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
C1RA_MOUSE
|
||||||
| NC score | 0.052508 (rank : 82) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
NETO1_HUMAN
|
||||||
| NC score | 0.052193 (rank : 83) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
KREM1_MOUSE
|
||||||
| NC score | 0.051867 (rank : 84) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
NETO1_MOUSE
|
||||||
| NC score | 0.051669 (rank : 85) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
TMPS7_MOUSE
|
||||||
| NC score | 0.051470 (rank : 86) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BIK6 | Gene names | Tmprss7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TMPS4_HUMAN
|
||||||
| NC score | 0.050309 (rank : 87) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRS4, Q5XKQ6, Q6UX37, Q9NZA5 | Gene names | TMPRSS4, TMPRSS3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Membrane-type serine protease 2) (MT-SP2). | |||||
|
TMPS2_MOUSE
|
||||||
| NC score | 0.049233 (rank : 88) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
SYNP2_MOUSE
|
||||||
| NC score | 0.036600 (rank : 89) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
CETP_HUMAN
|
||||||
| NC score | 0.029323 (rank : 90) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11597, Q13987, Q13988 | Gene names | CETP | |||
|
Domain Architecture |

|
|||||
| Description | Cholesteryl ester transfer protein precursor (Lipid transfer protein I). | |||||
|
HSH2D_MOUSE
|
||||||
| NC score | 0.026346 (rank : 91) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.022343 (rank : 92) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
AMPD3_MOUSE
|
||||||
| NC score | 0.021827 (rank : 93) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08739, O88692 | Gene names | Ampd3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (AMP deaminase H-type) (Heart-type AMPD). | |||||
|
DMRTD_HUMAN
|
||||||
| NC score | 0.021017 (rank : 94) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
AMPD3_HUMAN
|
||||||
| NC score | 0.018481 (rank : 95) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01432 | Gene names | AMPD3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (Erythrocyte AMP deaminase). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.017595 (rank : 96) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
CENPC_HUMAN
|
||||||
| NC score | 0.015274 (rank : 97) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.013648 (rank : 98) | θ value | 0.0961366 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
NUPL2_MOUSE
|
||||||
| NC score | 0.013646 (rank : 99) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.011596 (rank : 100) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.010637 (rank : 101) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.010636 (rank : 102) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
EFHD1_MOUSE
|
||||||
| NC score | 0.010519 (rank : 103) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D4J1, Q543U4 | Gene names | Efhd1, Sws2 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
ADA12_HUMAN
|
||||||
| NC score | 0.009652 (rank : 104) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43184, O60470 | Gene names | ADAM12, MLTN | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
GRN_HUMAN
|
||||||
| NC score | 0.009338 (rank : 105) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.009120 (rank : 106) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.008371 (rank : 107) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.008272 (rank : 108) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.007420 (rank : 109) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.006906 (rank : 110) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
PAX2_MOUSE
|
||||||
| NC score | 0.006855 (rank : 111) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32114 | Gene names | Pax2, Pax-2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-2. | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.005402 (rank : 112) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.004296 (rank : 113) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.003315 (rank : 114) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.003109 (rank : 115) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
HXC10_HUMAN
|
||||||
| NC score | 0.001819 (rank : 116) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYD6, O15219, O15220, Q9BVD5 | Gene names | HOXC10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C10. | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.000626 (rank : 117) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
SPEG_MOUSE
|
||||||
| NC score | -0.002405 (rank : 118) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ZN366_HUMAN
|
||||||
| NC score | -0.003206 (rank : 119) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N895, Q7RTV4 | Gene names | ZNF366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 366. | |||||
|
MARK4_HUMAN
|
||||||
| NC score | -0.003366 (rank : 120) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96L34, Q8NG37, Q96JG7, Q96SQ2, Q9BYD8 | Gene names | MARK4, KIAA1860, MARKL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 4 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase-like 1). | |||||
|
ZN606_HUMAN
|
||||||
| NC score | -0.003670 (rank : 121) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXB4, Q8NE04, Q96JH5 | Gene names | ZNF606, KIAA1852, ZNF328 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 606 (Zinc finger protein 328). | |||||